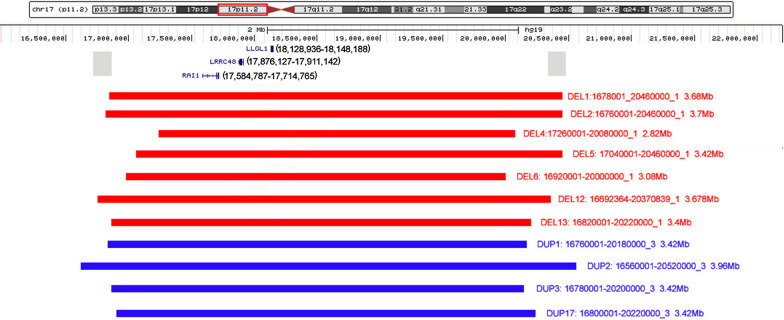
Fig. 3.
Chromosome 17p11.2 imbalance detected by NGS based CNV-seq. The region of 17p11.2 is framed by red line. Locations of RAI1, LRRC48 (DRC3) and LLGL1 genes are marked according to UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly (https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr17%3A16000001%2D22200000&hgsid=1093737035_AV6lvLz0lLX1TTtV27tDLC7HUtmy). The gray squares represent the locations of low copy repeats that mediate the common recurrent deletion/duplication, which is about 3.7 Mb. Copy number deletion and duplication are represented in red and blue bar, separately. The size and breakpoint of deletion and duplication are listed on the right of the bar. DEL1 represents deletion case No.1 (sorted in Table 1); DUP1 represents duplication case No.1 (sorted in Table 2), and the like. DEL13 represents the fetus with deletion; DUP17 represents the fetus with duplication. NGS: next generation sequencing. CNV: copy number variation