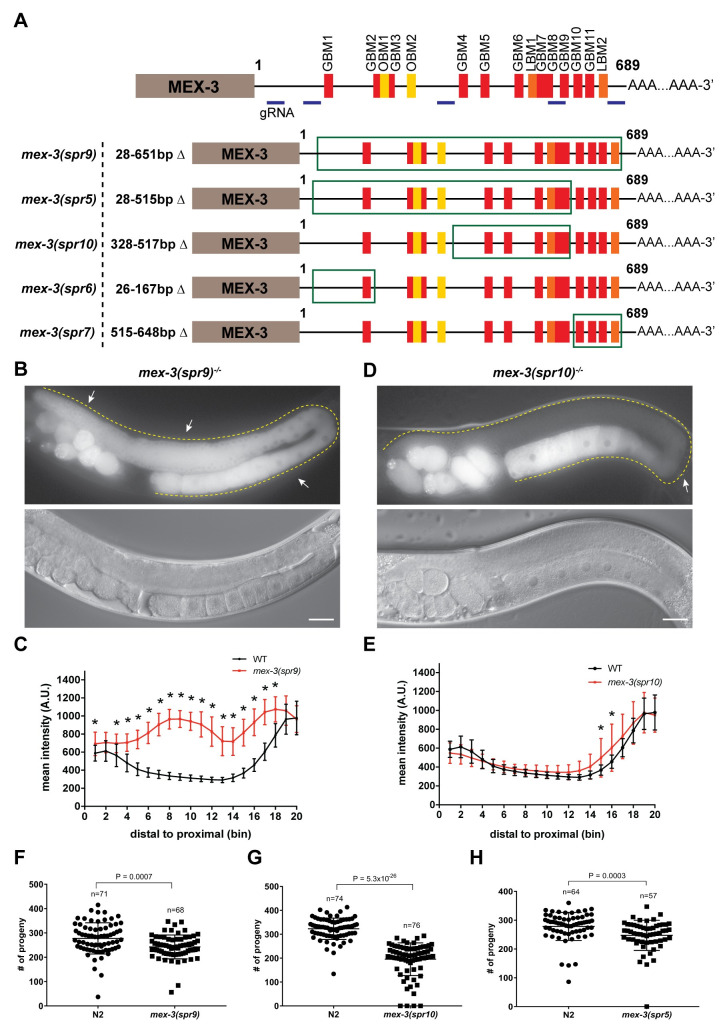
Fig 2. Deleting majority of the endogenous mex-3 3´UTR alters MEX-3 expression pattern and reduces fecundity.
(A) A schematic representing the 3´UTR deletions made in the wild type GFP::MEX-3 strain. The region deleted in each mutant is highlighted by the green rectangle. The locations of the gRNAs used in CRISPR/Cas9 mutagenesis are highlighted. (B) DIC and fluorescence images of the germline of the homozygous mex-3(spr9) mutant animals. (C) quantitative analysis of the fluorescence intensity in the mex-3(spr9) mutant animals compared to that of wild type GFP::MEX-3 (n = 23). (D) DIC and fluorescence images of the germline of the homozygous mex-3(spr10) mutant animals. (E) quantitative analysis of the fluorescence intensity in the mex-3(spr10) mutant animals compared to that of wild type GFP::MEX-3 (n = 17). (F) brood size assay of homozygous mex-3(spr9) mutant animals at 20°C. Each dot represents the brood size of an individual animal. Data from three biological replicates are shown in the graph. P-values are from a Kolmogorov-Smirnov test. (G) brood size assay of homozygous mex-3(spr10) mutant animals at 20°C. (H) brood size assay of mex-3(spr5) mutant animals at 20°C. All p-values for panels C and E are reported in S4 Table. All images taken at 40x magnification. Scale bar = 30 μm.