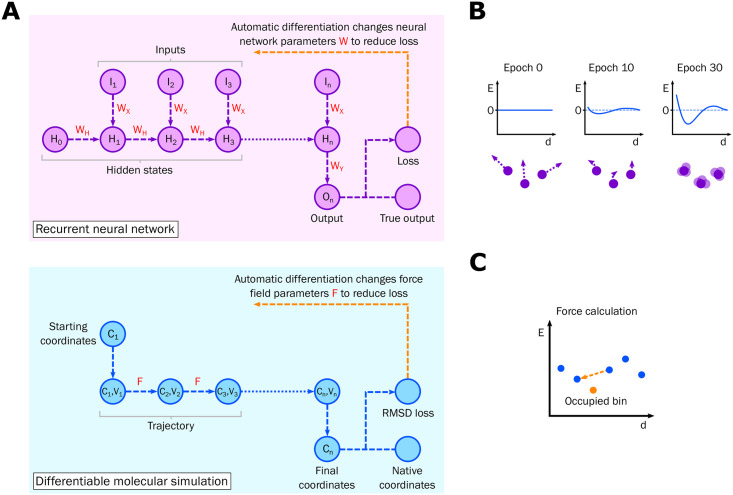
Fig 1. Differentiable molecular simulation.
(A) The analogy between a RNN and DMS. Learnable parameters are shown in red. The same parameters are used at each step. There are many variants of RNNs; the architecture shown here has a single output for a variable length input, which could for example represent sentiment classification of a series of input words. (B) Learning the potential. A representation of a component of the potential is shown with energy E plotted against inter-atomic distance d. At the start of training (epoch 0) the potential is flat and the atoms deform according to their starting velocities. During training the potential learns to stabilise the native structures of the training set. (C) Force calculation from the potential. Adjacent bins to the occupied bin are used to derive the force using finite differences. In this case the force acts to reduce the distance d. See the methods for more details.