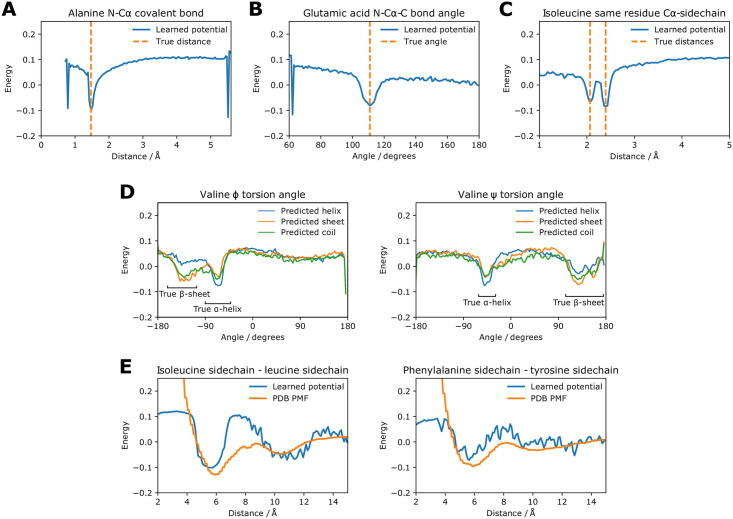
Fig 2. Components of the learned coarse-grained potential compared to true values.
The energy scale is the same for each plot. Each individual potential consists of discrete energy values for 140 evenly-spaced bins. (A) Distance potential between N and Cα in the same alanine residue, i.e. a covalent bond. (B) Bond angle potential for the N-Cα-C angle in glutamic acid. (C) Distance potential between Cα and the sidechain centroid on the same residue in isoleucine. (D) Torsion angle potentials in valine. There are different potentials for residues predicted as α-helical, β-sheet and coiled. The true ranges of α-helices and β-sheets are shown. (E) Distance potentials between sidechains for two pairs of amino acids. A PMF calculated from the PDB is also shown for comparison.