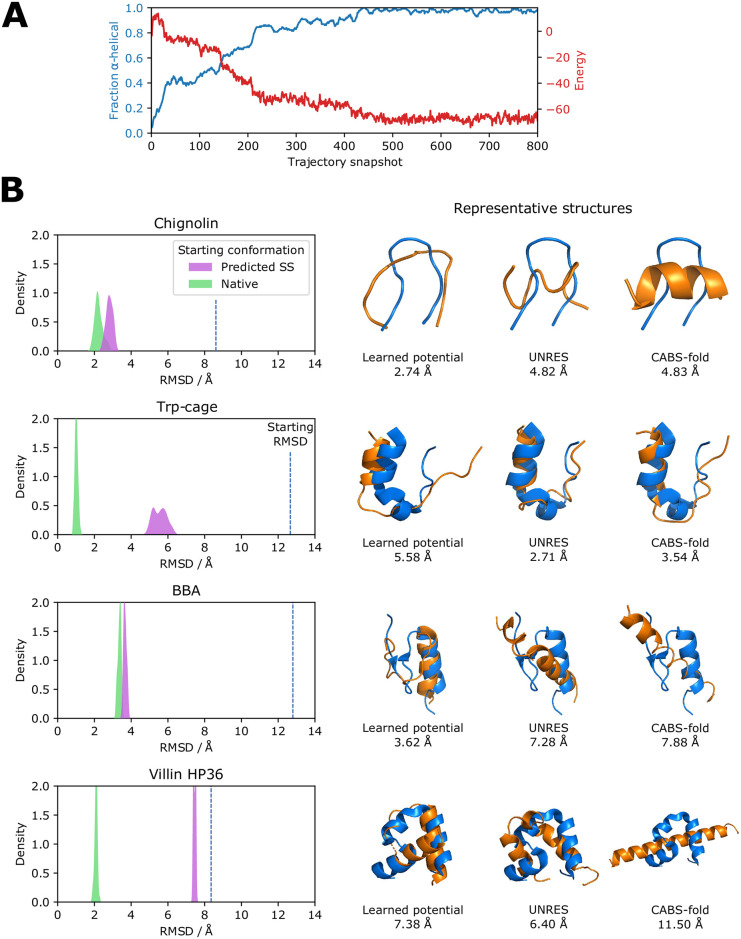
Fig 4. Folding small proteins and peptides.
(A) The (AAQAA)3 repeat peptide folds from a random starting conformation into an α-helix over 12m steps. The α-helical fraction and energy in the learned potential are shown with a snapshot taken every 15,000 steps. (B) Cα RMSD distributions from simulations of 3m steps for 4 proteins starting from predicted secondary structure and native conformations. An initial burn-in period of 9m steps was discarded. The starting Cα RMSD for the predicted secondary structure conformation is also shown. A representative structure found with MDAnalysis is shown (orange) along with models generated from the web servers of UNRES and CABS-fold. The Cα RMSD to the native structure (blue) is given for each model.