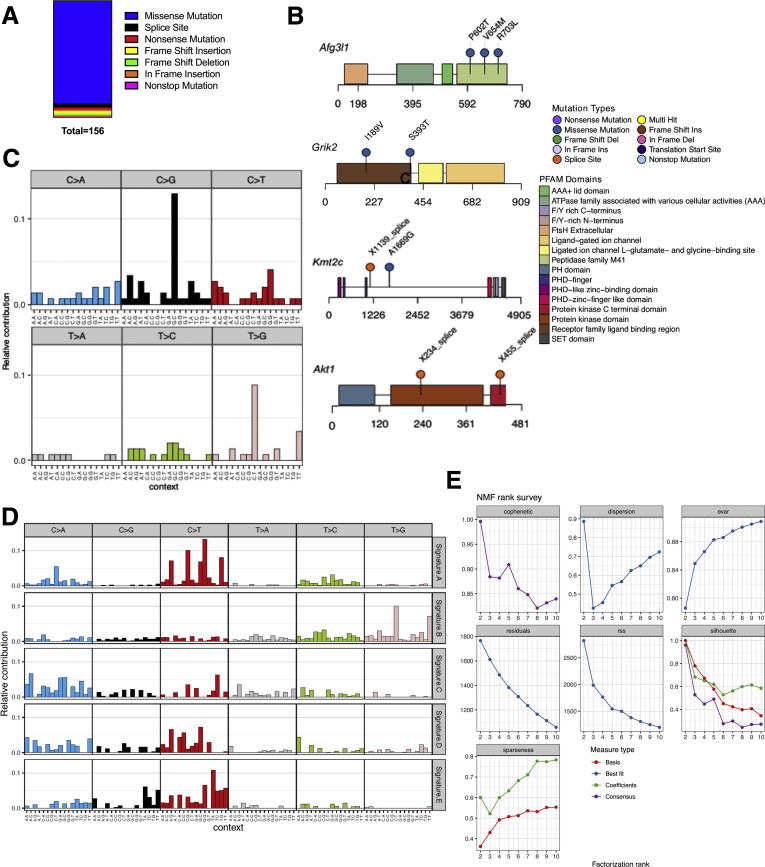
Figure 6.
WES of SB1 tumor cells. (A) Summary of mutation number and type from WES of the SB1 cell line. (B) Lollipop plots for Afg3I1, Grik2, Kmt2C, and Akt1 showing a schematic of each protein with protein domains from PFAM (protein family database) highlighted in colored blocks. Location of mutations is annotated with pins along the length of the protein. (C) Single-base substitution (SBS) signature of SB1 cell line. Each of the 6 mutation classes, as indicated at the top of the panel, is separated into 16 categories based on the identity of the preceding and following nucleotide as shown below. (D) SBS signatures for the 5 consensus signatures (Signature A–E) derived from TCGA-CHOL, arranged in rows. (E) Diagnostic plots for NMF rank survey. Cophentic correlation (top-left) was used to determine the optimal rank (five). AAA, ATPases associated woth diverse cellular activities; ATPase, adenosine triphosphatase; Del, deletion; F/Y, phenylalanin/tyrosine; NMF, non-negative matrix factorisation; PHD, plant homeodomain; SET, suvar3-9, enhancer-of-zeste and trithorax.