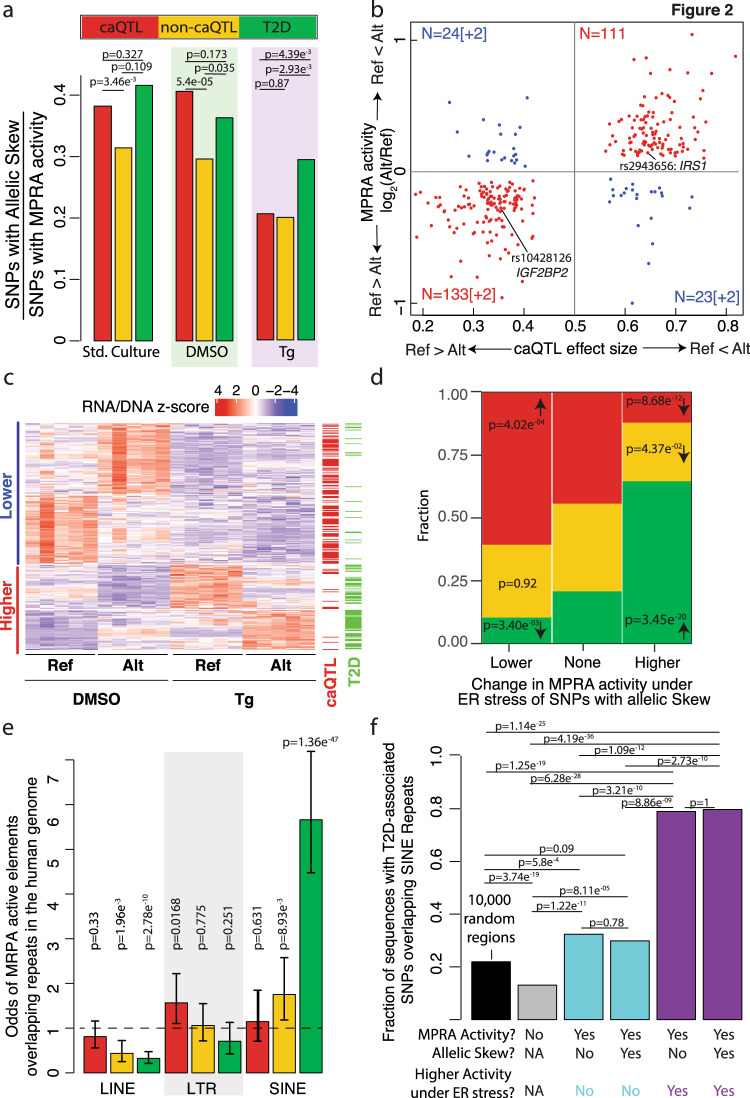
Fig. 2. Identification of β cell transcription-modulating allele.
a Fraction of caQTL, non-caQTL, and T2D SNP alleles that significantly alter MPRA activity in standard culture, DMSO, and Tg conditions. b Correlation between allelic effects on in vivo islet chromatin accessibility (x-axis) and MPRA activity (y-axis) for MPRA active elements containing islet caQTL SNPs. Quadrants 1 and 3 (red) contain SNPs where the allelic effects on chromatin accessibility and MPRA activity were concordant. The number of SNPs in each quadrant is indicated; the number of SNPs for which the relative MPRA activity lies outside the y-axis boundaries are indicated in brackets. Ref = hg19 reference allele; Alt = alternate allele. c Heatmap of elements containing SNPs that significantly alter MPRA activity for which overall MPRA activity is higher (red) or lower (blue) in Tg-treated vs. DMSO control-treated cells. Scale bar on top indicates z-scores obtained by centering and scaling the normalized RNA/DNA ratios for the reference and alternate alleles (5 replicates each per condition). Annotations on the right indicate whether the element (row) contains a caQTL and/or T2D-associated SNP. Note that the majority of elements with higher MPRA activity under ER stress contain T2D-associated SNPs. d Proportion of elements containing caQTL (red), non-caQTL (yellow), or T2D-associated (green) SNPs with allelic effects on MPRA activity (under any condition) that show lower, no change, or higher MPRA activity under ER stress conditions compared to DMSO control. A significant proportion of caQTL SNPs with allelic skew showed lower MPRA activity under ER stress, while a significant proportion of T2D-associated SNPs showed higher activity in this condition. ***p < 0.001; **p < 0.01; and *p < 0.05, Fisher’s exact test comparing elements with higher or lower MPRA activity under ER stress to those showing no change. e Odds of MPRA active elements (≥1 allele; any experimental condition) containing caQTL, non-caQTL, or T2D-associated SNPs for overlapping long interspersed nuclear element (LINE), long terminal repeat (LTR), or short interspersed nuclear element (SINE) repetitive elements. Error bars indicate the 95% confidence interval of the odds ratio estimates. f Fraction of T2D SNP-containing elements or 10,000 randomly selected 200 bp genomic regions (black bar) overlapping SINE repeats in the human genome. T2D SNP-containing elements are grouped based on whether they show (i) MPRA activity, (ii) allelic skew in MPRA activity, and/or (iii) higher MPRA activity under ER stress. A significantly higher fraction of T2D SNP-containing elements with higher activity under ER stress overlap SINEs compared to those without higher activity under ER stress. ***p < 0.001; **p < 0.01; and *p < 0.05 using two-sided Fisher’s exact test (corrected for multiple testing) in panels (a), (d), (e), and (f).