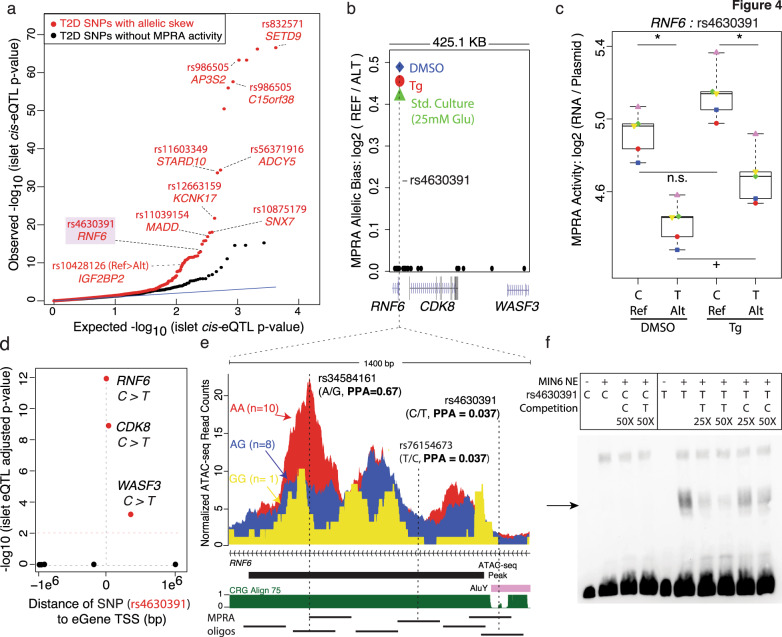
Fig. 4. MPRA identifies putative T2D causal SNPs altering islet expression (eQTL) and chromatin accessibility (caQTL).
a Quantile-quantile plot of observed (y-axis) vs. expected (x-axis) islet eQTL p-values from the InsPIRE Consortium for T2D SNPs, categorized based on whether ≥1 SNPs in LD had allelic skew (red dots), or 0 SNPs in LD had significant MPRA activity (black dots). b Plot of genome location of 27 SNPs in high LD (r2 ≥ 0.8) with the T2D-associated index SNP rs10507349 tested with MPRA (RNF6 locus). Allelic effects on MPRA activity were detected for rs4630391 in the same direction across all three experimental conditions. c MPRA activity for the reference (‘C’) and alternate (‘T’) alleles of rs4630391 in DMSO and Tg experimental conditions. Each of the five paired biological replicates for each MPRA experiment are indicated by distinct shapes and colors. * indicates significant (FDR < 10%) allelic effects on MPRA activity; + or n.s. indicates significant (FDR < 1%) or not significant changes in MPRA activity in each condition, respectively. Box plots display the minimum, maximum, median, first quartile and third quartile of each data set. d Plot of InsPIRE Consortium islet eQTL association p-values between rs4630391 genotypes and expression of genes ± 1 megabase. e Magnified view of genomic region containing the MPRA-nominated functional T2D SNP (rs4630391) and lead islet caQTL SNP (rs34584161). Credible set SNP genetic posterior probabilities of association (PPA) are included for SNPs for which they were reported4. Normalized ATAC-seq reads were higher for islets from donors with rs34584161 AA homozygous genotypes (red; n = 10 donors) than those from AG heterozygous (blue; n = 8) or GG homozygous (yellow; n = 1) genotypes. ‘CRG Align 75’ indicates mappability of 75-mer sequences to the hg19 reference genome. Thick black bar indicates ATAC-seq peak in human islets. Thin black lines below indicate the MPRA oligos tested in this region. f Allele-specific binding of sequences containing reference and alternate alleles for rs4630391. EMSA with biotin-labeled probes containing rs4630391 C or T alleles were incubated with MIN6 nuclear extract. The arrow indicates allele-specific nuclear factor binding to the T allele, which was more efficiently competed with unlabeled T probe than unlabeled C probe.