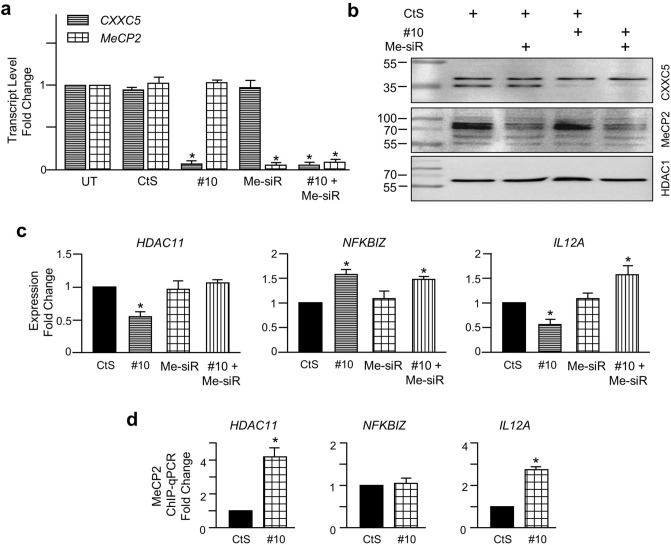
Figure 8.
Assessing the interplay between CXXC5 and MeCP2 in CXXC5 target gene expressions. (a) MCF7 cells were untransfected (UT), or were transiently transfected with CtS, siRNA specific for CXXC5 (#10), and/or siRNA pool for MeCP2 (Me-SiR) for 48 h. To equalize the total amount of siRNA (20 nM) used in co-transfection experiments, 10 nM gene-specific siRNA was used together with 10 nM CtS. cDNA generated from total RNA were subjected to qPCR. The RPLP0 expression was used for normalization. Results, the mean ± S.E. of three independent determinations, depict fold change in comparison with transcript levels of CXXC5 or MeCP2 of UT, which were set to 1. The asterisk indicates a significant difference. (b) Nuclear extracts of transfected cells were also subjected to WB using the CXXC5 or MeCP2 antibody. HDAC1 was probed with an HDAC1-specific antibody. MMs in kDa are indicated. Uncropped blots are presented in Supplementary Information, Figure S11. (c) To assess the effect of reduction in CXXC5 and/or MeCP2 levels on gene expressions, MCF7 cells were transfected with CtS, #10, and/or Me-SiR as indicated for 48 h. cDNAs were then subjected to qPCR using primer sets specific to target genes. RPLP0 expressions were used for normalization. Results as fold change in gene expressions compared to those observed in CtS transfected cells are the mean ± S.E. of three independent determinations. Asterisks indicate significant differences. (d) To assess whether alterations in CXXC5 levels affect the MeCP2 loading on target promoters, MCF7 cells transfected with CtS (10 nM) or #10 (10 nM) for 48 h were subjected to ChIP using IgG or a MeCP2 antibody. Recovered DNAs were subjected to qPCR using primer sets for target gene promoters. Results, normalized to IgG, depict fold changes compared to CtS, which was set to 1.