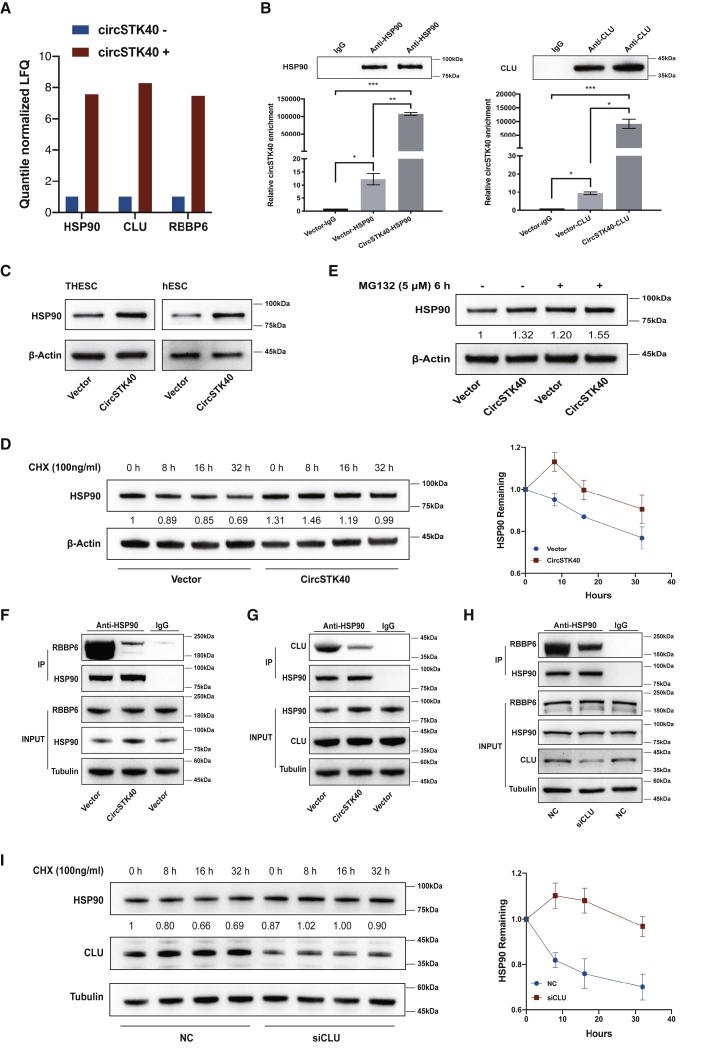
Figure 3.
circSTK40 blocks the interaction between HSP90 and CLU, thus reducing the proteasomal degradation of HSP90
(A) Fold changes of HSP90 and CLU detected through a liquid chromatography-based mass spectrometry analysis after circSTK40 pull-down assays. The antisense transcript of circSTK40 was used as the negative probe. (B) Enrichment of circSTK40 in the RNA immunoprecipitates of anti-HSP90 (left) or anti-CLU (right) antibodies in day 4-induced THESCs infected with a control vector or a circSTK40-overexpression adenovirus (n = 3). (C) The protein levels of HSP90 in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs. (D) The protein levels of HSP90 at 0, 8, 16, and 32 h after treatment with CHX (a protein synthesis inhibitor, 100 ng/mL) in day 4-induced, circSTK40-overexpressing THESCs (left). The results of relative quantification are expressed as the mean ± SEM (n = 3) and the degradation curves (right). (E) The protein levels of HSP90 in day 4-induced, circSTK40-overexpressing THESCs after treatment with or without MG132 (a proteasome inhibitor, 5 μM) for 6 h. (F) The association between HSP90 and RBBP6 detected using coIP with anti-HSP90 antibody in day 4-induced, circSTK40-overexpressing THESCs. (G) The association between HSP90 and CLU detected using coIP with anti-HSP90 antibody in day 4-induced, circSTK40-overexpressing THESCs. (H) The association between HSP90 and RBBP6 detected using coIP with anti-HSP90 antibody in day 4-induced THESCs after CLU knockdown with siRNA. (I) The protein levels of HSP90 at 0, 8, 16, and 32 h after treatment with CHX (100 ng/mL) in day 4-induced THESCs after CLU knockdown with siRNA (left). The results of relative quantification are expressed as the mean ± SEM (n = 3) and the degradation curves (right). Data are presented as the mean ± SEM. Student’s t test was used for determining the differences between two independent samples; an ANOVA was used for determining the overall differences within multiple samples. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; n.s., non-significant.