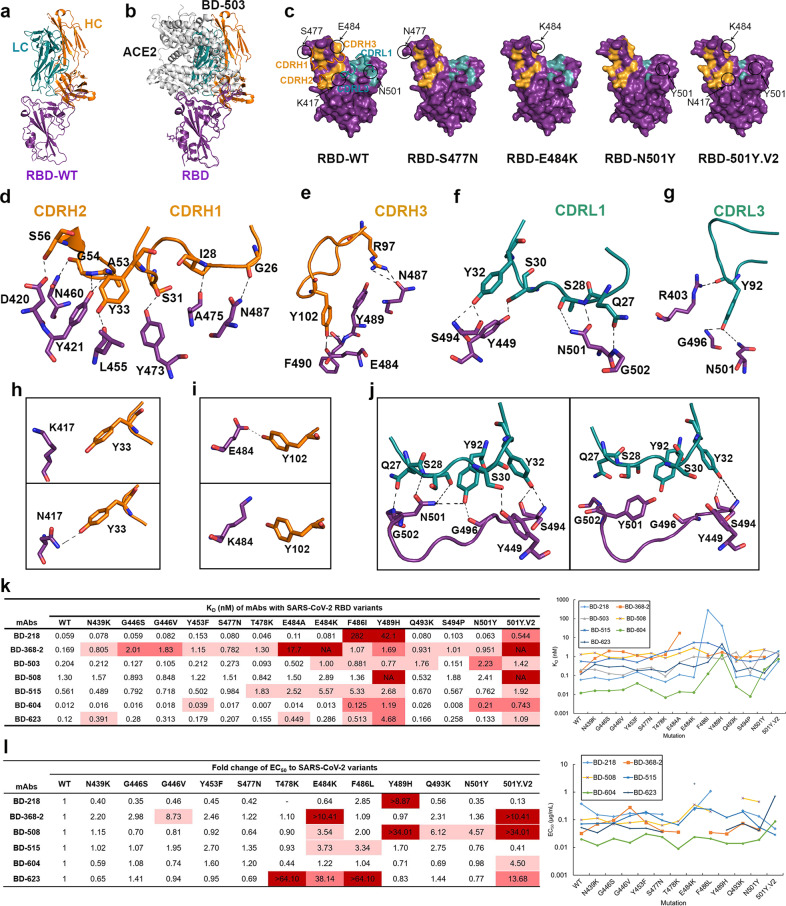
Fig. 1. Interaction of SARS-CoV-2 RBD variants with neutralizing antibodies.
a Crystal structure of BD-503 in complex with RBD. b BD-503 clashed with ACE2 binding to SARS-CoV-2 RBD, and therefore interfered with the interaction between RBD and ACE2. c Epitopes of BD-503 with SARS-CoV-2 RBD, RBD-S477N, RBD-E484K, RBD-N501Y and RBD-501Y.V2. CDRH1, CDRH2, CDRH3, CDRL1, and CDRL3 were involved in the interaction. The RBD is shown in a surface view. d–g Detailed contacts between BD-503 and SARS-CoV-2 RBD. h Y33 in the heavy chain of BD-503 can recognize K417N mutation site in RBD. i The E484K mutation in RBD broke the interaction between N484 and Y102 in the heavy chain of BD-503. j The N501Y mutation in RBD disrupted the interaction between N501 and BD-503 and broke the contact of G502 in RBD and Q27 in BD-503 light chain, G496 in RBD and Y92 in BD-503 light chain, Y449 in RBD and S30 in BD-503 light chain. k KDs of neutralizing antibodies binding to SARS-CoV-2 RBD and to naturally occurring variants as measured by SPR. Red indicates major fold-change larger than 100-fold. Rose indicates fold-change between 10- and 100-fold. Pink indicates minor fold-change between 3- and 10-fold. l EC50s and fold-changes of NAbs against pseudovirus carrying SARS-CoV-2 RBD mutations. Red indicates major fold-change that cannot be obtained from the assay. Rose indicates fold-change between 10- and 100-fold. Pink indicates minor fold-change between 3- and 10-fold.