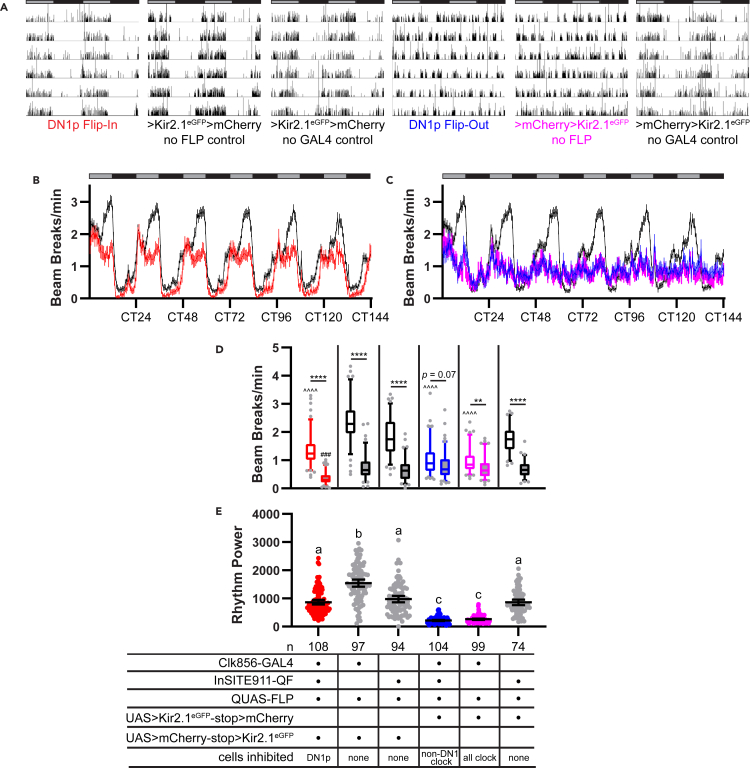
Figure 3.
Sustained free-running rest:activity rhythms following selective and comprehensive expression of Kir2.1eGFP in DN1p cells
(A) Representative single-fly activity records are shown for the genotypes listed. Activity in infrared beam breaks/min is plotted for each minute over 6 d in DD conditions. Data are double plotted, with 48 hr on each line and the second 24 hr replotted at the start of the next line. Gray and black bars above each plot represent subjective day and night, respectively.
(B) Mean number of DAM beam breaks/min (±95% confidence interval) is plotted over 6 days in DD conditions for DN1p Flip-In flies (red) compared with control flies (black).
(C) Mean DAM beam breaks/min is plotted as in (B) for DN1p Flip-Out flies (blue), flies in which all Clk856+ clock neurons are silenced with Kir2.1eGFP (pink), and control flies (black). The same control data are graphed in (B) and (C) and consist of a combination of 3 control lines lacking Kir2.1eGFP expression (see STAR Methods). Gray and black bars indicate subjective day and night, respectively.
(D) Box and whisker plots of mean DAM beam breaks/min over the 12-hr subjective day (open boxes) and 12-hr subjective night (filled boxes) are shown for the indicated genotypes. Box extends from 25th to 75th percentiles; whiskers extend to 5th and 95th percentiles. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, within genotype comparison between subjective day and night activity; ˆˆˆˆp < 0.0001 compared to subjective day activity of all 3 control lines; ##p < 0.01 compared with subjective night activity of all three control lines, Tukey’s multiple comparisons test following two-way ANOVA.
(E) Rest:activity rhythm power over 6 days in DD is displayed for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. Different letters indicate significant difference (p < 0.05), Tukey’s multiple comparisons test following one-way ANOVA. The same letter indicates no significant different (p > 0.05).