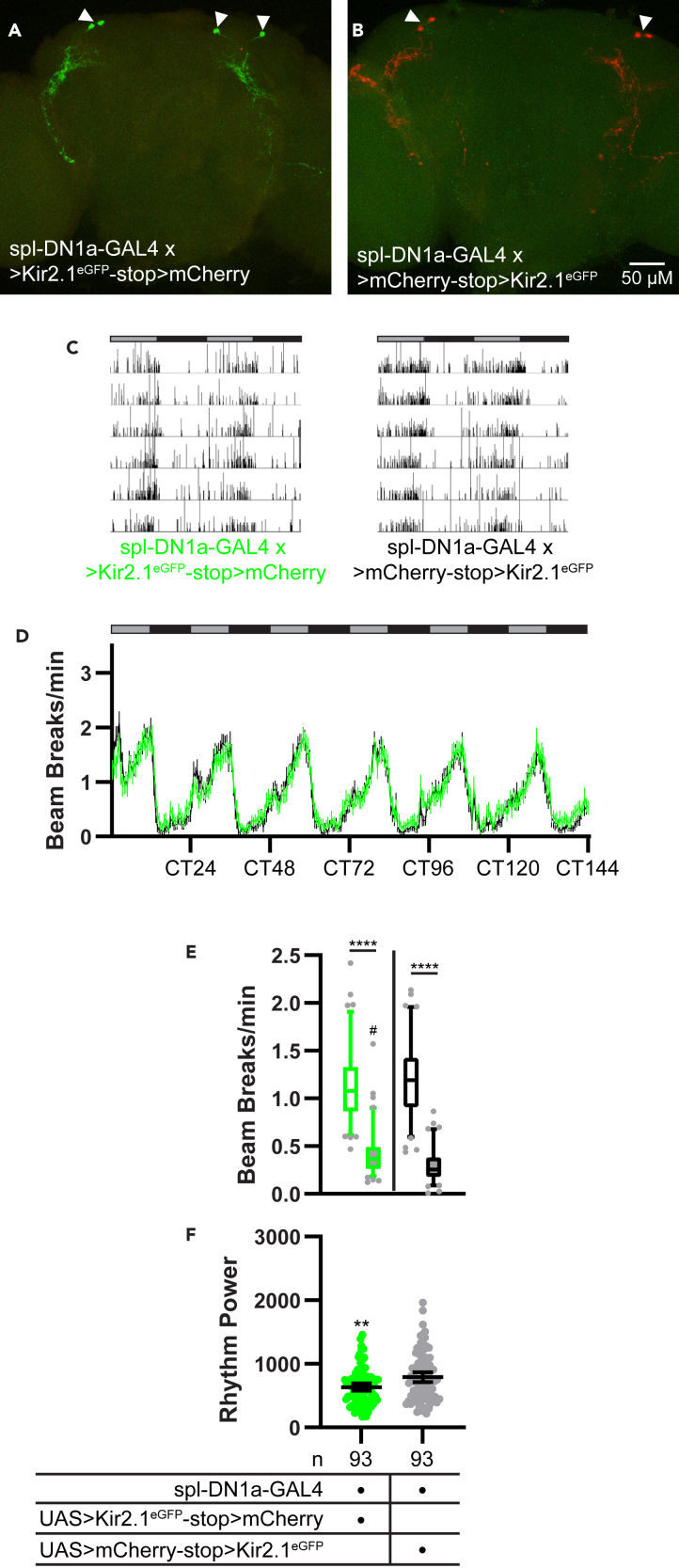
Figure 7.
Minor effects of DN1a silencing on free-running rest:activity rhythms
(A and B) Representative maximum projection confocal images of brains from flies in which spl-DN1a-GAL4 was used to drive UAS>Kir2.1eGFP-stop>mCherry (A) or UAS>mCherry-stop>Kir2.1eGFP (B). Brains were co-stained for Kir2.1eGFP (green) and mCherry (red) immunofluorescence. Arrowheads indicate DN1a cells. Note that expression was restricted to 2 DN1a cells per hemisphere with this split GAL4 line.
(C) Representative single-fly activity records are shown for the genotypes listed. Activity in infrared beam breaks/min is plotted for each minute over 6 d in DD conditions.
(D) Mean number of DAM beam breaks/min (±95% confidence interval) is plotted over 6 days in DD conditions for flies with spl-DN1-GAL4-driven Kir2.1eGFP (green) and control flies with spl-DN1a-GAL4-driven mCherry (black). Gray and black bars indicate subjective day and night, respectively.
(E) Box and whisker plots of mean DAM beam breaks/min over the 12-hr subjective day (open boxes) and 12-hr subjective night (filled boxes) are shown for the indicated genotypes. Box extends from 25th to 75th percentiles; whiskers extend to 5th and 95th percentiles. ∗∗∗∗p < 0.0001, within genotype comparison between subjective day and night activity; #p < 0.05 compared with subjective night activity of control flies; Tukey’s multiple comparisons test following two-way ANOVA.
(F) Rest:activity rhythm power over 6 days in DD is plotted for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. ∗∗p < 0.01, t test.