Figure 1.
Necessity of a dual-mutation base editor with minimal PAM restriction for simulating MNVs
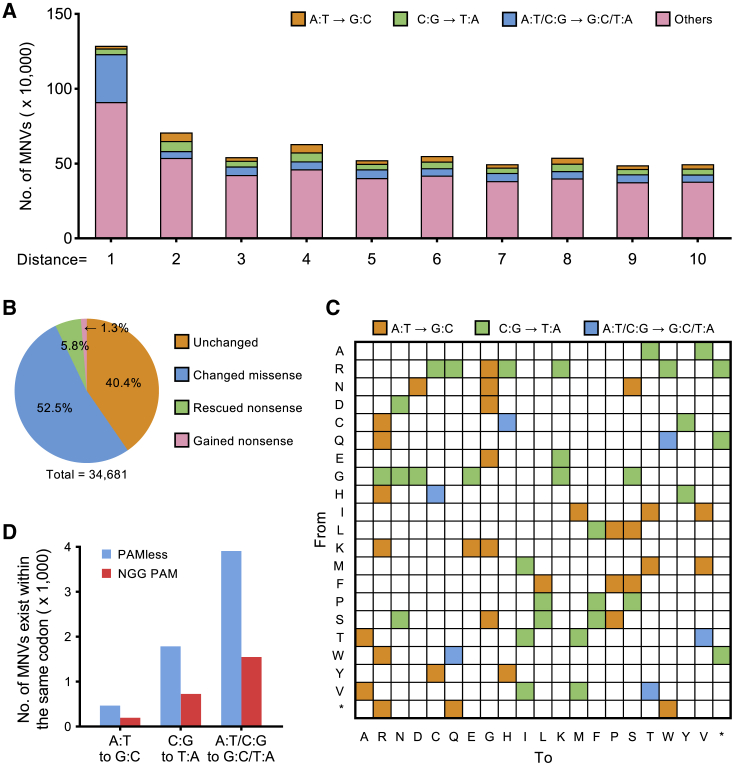
(A) Substitution pattern of MNVs from gnomAD. The x coordinate is the distance between two SNVs. (B) Differences in functional impacts on the protein between MNVs and individual SNPs. (C) Heatmap showing the amino acid changes that can be simulated by the dual-mutation base editor. (D) The PAM-flexible dual-mutation base editor has a broadened scope to target MNVs within the same codon. A broadened editing window (positions 3–10) was used to calculate the targeting scope.