Summary
Broadly neutralizing antibodies (bnAbs) against HIV-1 are frequently associated with the presence of autoreactivity/polyreactivity, a property that can limit their use as therapeutic agents. The bnAb 4E10, targeting the conserved Membrane proximal external region (MPER) of HIV-1, displays almost pan-neutralizing activity across globally circulating HIV-1 strains but exhibits nonspecific off-target interactions with lipid membranes. The hydrophobic apex of the third complementarity-determining region of the heavy chain (CDRH3) loop, which is essential for viral neutralization, critically contributes to this detrimental effect. Here, we have replaced the aromatic/hydrophobic residues from the apex of the CDRH3 of 4E10 with a single aromatic molecule through chemical modification to generate a variant that preserves the neutralization potency and breadth of 4E10 but with reduced autoreactivity. Collectively, our study suggests that the localized accumulation of aromaticity by chemical modification provides a pathway to ameliorate the adverse effects triggered by the CDRH3 of anti-HIV-1 MPER bnAbs.
Subject areas: immunology, virology
Graphical abstract
Highlights
-
•
Aromatic grafting is employed to improve functionality of the HIV antibody 4E10
-
•
Engineering the CDRH3 loop slashes its polyreactivity profile but also its potency
-
•
Site-specific chemical modification rescues the activity of the engineered antibody
-
•
Collectively, this procedure mitigates the polyreactivity of an MPER antibody
Immunology; Virology
Introduction
Antibodies, and in particular broadly neutralizing antibodies (bnAbs), have emerged as promising therapeutic options against infectious diseases because of their ability to selectively neutralize target pathogens and to flag them for elimination (Walker and Burton, 2018). Following the advent of single B cell antibody cloning and high-throughput neutralization assays, the collection of available HIV-bnAbs has dramatically expanded over the last decade (Klein et al., 2013; Kwong and Mascola, 2012; Sok and Burton, 2018).
Among the anti-HIV-1 bnAbs isolated so far, those targeting C-terminal residues of the conserved membrane-proximal external region (MPER) of the gp41 subunit consistently display the broadest levels of viral neutralization (Huang et al., 2012; Krebs et al., 2019; Kunert et al., 2004; Pinto et al., 2019; Stiegler et al., 2001; Williams et al., 2017; Zhang et al., 2019). That is, Abs targeting this highly conserved HIV-1 Env sequence block viral infection of a wide range of circulating HIV-1 isolates. These bnAbs, when delivered passively in animal models, prevent or suppress viral infection (Asokan et al., 2015; Ferrantelli et al., 2003; Hessell et al., 2010; Huang et al., 2016; Pegu et al., 2014; Stiegler and Katinger, 2003). However, MPER bnAbs commonly display polyreactivity/autoreactivity, particularly against lipid antigens (Haynes et al., 2005), a feature that has cautioned against their clinical use in passive therapies (Alving, 2008; Haynes et al., 2005).
Autoreactivity/polyreactivity of 4E10 has been extensively characterized (Alam et al., 2007; Dennison et al., 2009; Haynes et al., 2005; Matyas et al., 2009). In fact, the mechanism by which 4E10 binds to the MPER helix with high affinity requires the insertion of the complementarity-determining region of the heavy chain (CDRH3) loop into the membrane (Carravilla et al., 2020; Irimia et al., 2016; Rantalainen et al., 2020; Rujas et al., 2017), explaining the collateral binding to membrane lipids (Haynes et al., 2010). Consistent with this observation, substitutions and deletions of the hydrophobic residues encoded in the key D-gene severely interfere with binding to native virions and viral neutralization (Alam et al., 2009; Carravilla et al., 2019; Rujas et al., 2015; Scherer et al., 2010).
Here, we have re-engineered the CDRH3 loop of the human bnAb 4E10 (100WGWL100c) to minimize off-target interactions with the lipid membrane while preserving its neutralization potency. Specifically, we have removed the hydrophobic residues at the apex of the CDRH3 and replaced them by a polycyclic aromatic compound (pyrene) previously described to improve antibody binding to native MPER in HIV-1 virions (Rujas et al., 2020). This trimming-and-grafting strategy concentrated the hydrophobicity/aromaticity, otherwise spanned over several residues of the CDRH3, into a single location of the loop resulting in a re-engineered 4E10 antibody variant with favorable functional properties. In particular, the resulting 4E10 variant displayed the same neutralization potency and breadth as that of the wild-type antibody but with notably less autoreactivity against membranes and HEp-2 cells. Collectively, our data support chemical engineering of CDRH3 hydrophobicity as a strategy to improve the functional profile of Abs that have evolved to bind epitopes located in the proximity of membrane interfaces.
Results
Removing hydrophobic residues from the tip of the CDRH3 loop decreases 4E10 autoreactivity
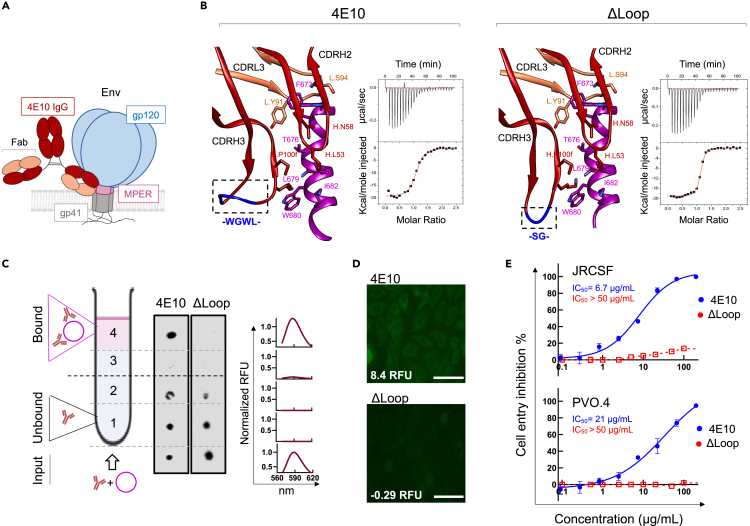
Binding of bnAb 4E10 to HIV-1 Env on the virus (Figure 1A) relies on the insertion of the aromatic-rich CDRH3 apex of sequence – WGWL – into the viral membrane. This interaction has been postulated to stabilize lateral antibody binding to the MPER helical epitope in the environment provided by the membrane interface (Irimia et al., 2016; Rujas et al., 2017; Scherer et al., 2010). However, the four residues – WGWL – are not required for specific binding to the MPER epitope in solution because their substitution with the shorter – SG – sequence yields the antibody variant termed ΔLoop, which recognizes the epitope-peptide with comparable affinity and structural interactions to that of the wild-type antibody (Figure 1B) (Rujas et al., 2015). However, since these hydrophobic residues are required to penetrate the lipid bilayer, the antigen-binding fragment (Fab) of the ΔLoop variant lost its ability to interact with membranes made of virus-like (VL) lipid mixtures emulating the virus envelope (Scherer et al., 2010) (Figure 1C). The inability to interact with lipids was also demonstrated by the absence of reactivity of ΔLoop with HEp-2 epithelial cells, which is a standard method to identify autoantibodies (Dellavance and Andrade, 2019; Haynes et al., 2005) (Figure 1D). Not surprisingly, the modification introduced in ΔLoop resulted in the abolition of HIV-1 neutralization activity at concentrations below 200 μg/mL (Figure 1E). Altogether, these data indicate that the redesign of the hydrophobic apex of the CDRH3 loop is a suitable a strategy to minimize off-target interactions of the antibody 4E10 with lipid membranes, provided that its manipulation does not interfere with its critical biological function.
Figure 1.
Effect of trimming the hydrophobic residues from the CDRH3 loop tip on the functional profile of 4E10
(A) Schematic representation of 4E10 antibody (dark red, heavy chain; orange, light chain) binding to the MPER region (pink) located in the gp41 subunit (gray) of the HIV-1 Env.
(B) MPER epitope binding by Fabs 4E10 (PDB: 4WY7) and its trimmed version ΔLoop (PDB: 5CIN) (left and right panels, respectively). Residues from the light (orange) and heavy (dark red) chain involved in epitope peptide binding with a buried surface area (BSA) > 35 Å are depicted with sticks. The peptide is shown in magenta. The binding isotherms to the epitope peptide (671NWFDITNWLWYIK683KKK) are also shown. Fitting to a one-site binding model yielded binding constants (KD-s) of 49 nM and 12 nM for the 4E10 and ΔLoop Fab, respectively.
(C) Partitioning to VL membranes was measured by a vesicle flotation assay, as schematically described in the left panel. The presence of Fabs in the different fractions was determined by Western-blot (middle panel). Representative Rhodamine fluorescence emission spectra displayed in the right panel evidenced the presence of the liposomes in the upper fractions of the 4E10 sample after flotation (pink traces). A similar Rhodamine signal distribution was observed for all samples measured in this study.
(D) Immunofluorescence staining against HEp-2 epithelial cells. Normalized RFU values are indicated (see Materials and Methods) Scale bars in the micrographs are 50 μm.
(E) Antiviral activity of Fabs 4E10 (blue) and ΔLoop (red) against JRCSF (tier 2) and PVO.4 (tier 3) Env-pseudotyped virions (mean values ±SD for two technical replicates). IC50 values are shown.
Covalent attachment of Fus4 to the trimmed loop restores neutralization breadth and potency without affecting structural stability
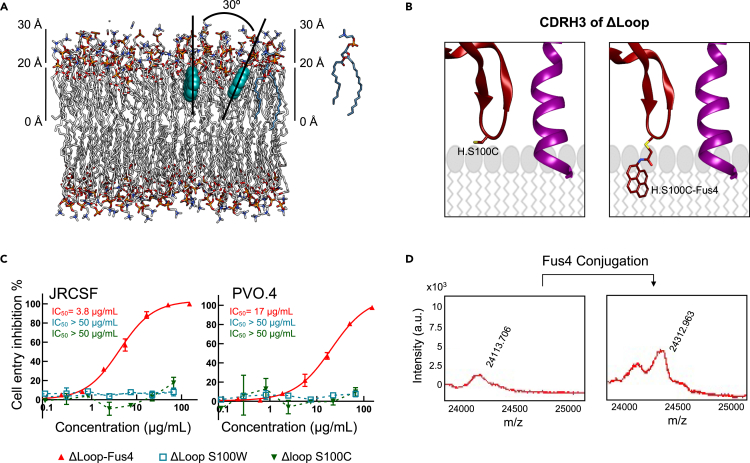
In view of the previous results, we aimed to rescue the biological function of the ΔLoop variant by restoring the hydrophobicity of the CDRH3 loop using a single molecule. Since the range of physicochemical properties of natural amino acids is limited, we considered the modification of the antibody with a chemical compound. To that end, we selected a pyrene-based acetamide compound with four fused aromatic rings that we termed Fus4. This is a highly hydrophobic/aromatic molecule that efficiently interacts with VL membranes (Rujas et al., 2020). Specifically, MD simulations revealed that Fus4 locates within one leaflet of the bilayer, with its center of mass submerged into the upper sections of the acyl chains, close to the interface, and with its longitudinal axis almost parallel to the membrane normal (Figure 2A) (Rujas et al., 2020).
Figure 2.
Fus4 engraftment onto the truncated CDRH3 restores antiviral activity of the Fab ΔLoop
(A) Depth of insertion and possible orientations adopted in the lipid bilayer by the aromatic compound Fus4. Lipids and Fus4 are depicted with sticks and spheres, respectively.
(B) Cartoon representation depicting a model of the Fus4 moiety being incorporated in the CDRH3 apex (orange). The epitope peptide is also shown (magenta).
(C) Cell-entry inhibition assays against JRCSF and PVO.4 PsV (mean values ±SD for two technical replicates) comparing unmodified ΔLoop H.S100C mutant (green), modified ΔLoop H.S100C-Fus4 (red), and H.S100W mutant (blue). To facilitate the comparison, IC50 values are indicated.
(D) MADi-TOF m/z plot before (left) and after (right) chemical modification. See also Figures S1 and S2; Table S1.
Owing to its large quadrupole moment, Fus4 was predicted to stabilize Fab interaction in the vicinity of the phosphoglycerol moieties more strongly than the natural Trp residue (containing two fused rings) (McDonald and Fleming, 2016; Yau et al., 1998). To verify this hypothesis, we modified the CDRH3 apex with a single Fus4 molecule (Figures 2B and 2D) resulting in the full recovery of the anti-viral potency of ΔLoop (Figure 2C). Notably, the presence of a Trp residue at the exact same position as that of Fus4 in ΔLoop (H.S100W mutant) did not restore the neutralization activity of the Fab (Figure 2C).
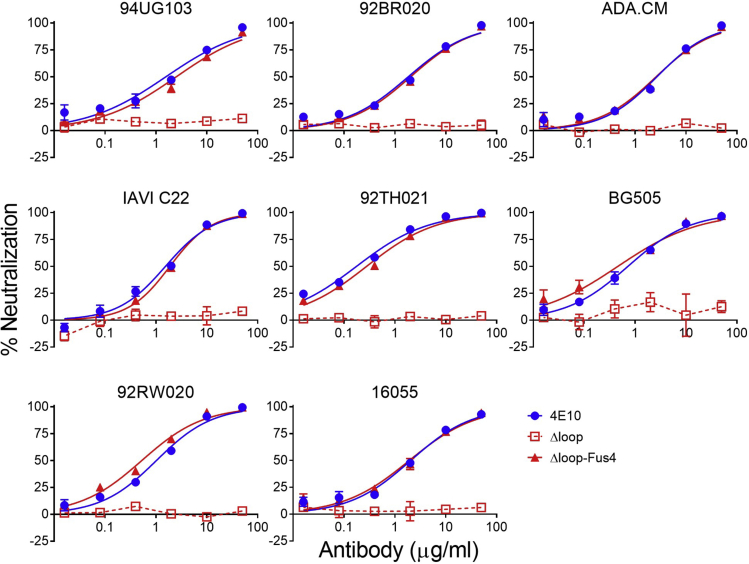
Neutralization potency and breadth of ΔLoop-Fus4 were further evaluated against a panel of HIV-1 isolates used previously as an indicator of cross-clade neutralization breadth (Simek et al., 2009) in a standardized in vitro TZM-bl neutralization assay (Sarzotti-Kelsoe et al., 2014) (Figure 3). The IC50 value and breadth of ΔLoop-Fus4 Fab was compared with the unmodified ΔLoop variant and to the wild-type 4E10 Fab. ΔLoop-Fus4 Fab displayed 100% breadth against this panel and neutralized each of the HIV-1 pseudoviruses (PsVs) with the same potency as the parental 4E10 Fab (Figure 3 and Table 1).
Figure 3.
Modified ΔLoop-Fus4 displays similar potency and breadth to that of parental 4E10
Representative neutralization titration curves of ΔLoop-Fus4 (solid red triangles, lines), parental 4E10 (solid blue circles, lines), and ΔLoop (empty red squares, dashed lines) against a panel of eight HIV-1 isolates previously described as an indicator of cross-clade neutralization breadth (Simek et al., 2009) (mean values ± SD for two technical replicates).
Table 1.
Fus4 conjugation-induced DLoop neutralization rescue measured against a variety of HIV isolates
| IC50 (μg/ml) |
|||||
|---|---|---|---|---|---|
| Virus | Clade | Tier | 4E10 | Δloop | Δloop-Fus4 |
| 94UG103 | A | 2 | 1.6 | >50 | 2.5 |
| 92BR020 | B | 2 | 1.8 | >50 | 2.0 |
| ADA.CM | B | 2 | 2.7 | >50 | 2.6 |
| IAVI C22 | C | 2 | 1.5 | >50 | 1.9 |
| 92TH021 | AE | 2 | 0.18 | >50 | 0.28 |
| BG505 | A | 2 | 0.72 | >50 | 0.46 |
| 92RW020 | A | 2 | 1.0 | >50 | 0.53 |
| 16055 | C | 2 | 2.1 | >50 | 1.9 |
X-ray crystallography and thermostability assays ruled out effects on the overall folding of the Fab induced by the chemical modification with Fus4 (Figures S1 and S2 and Table S1). Thus, together, the functional and structural data support the idea that synthetic compounds can provide beneficial biological properties without inducing structural alterations, and hence broaden the antibody engineering toolkit.
Fus4 engraftment promotes ΔLoop binding to the native antigen
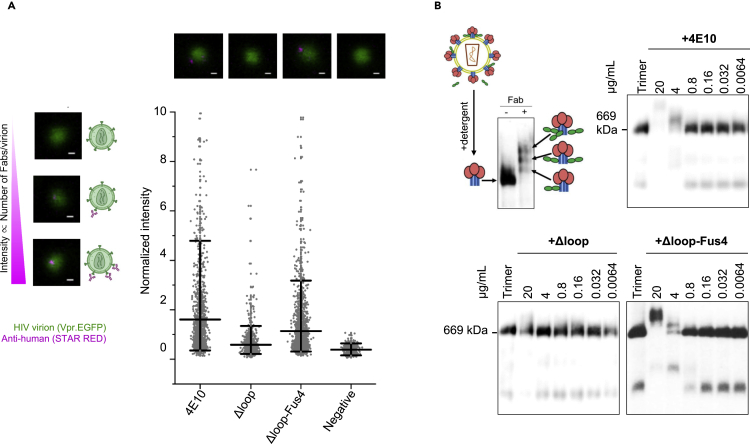
To gain further insights into the mechanism by which the neutralization activity of ΔLoop is recovered upon covalent addition of Fus4 to the CDRH3 tip, we measured the binding affinity of ΔLoop-Fus4 Fab to native Env in intact virions using super-resolution stimulated-emission depletion (STED) microscopy (Carravilla et al., 2019). To that purpose, we incubated EGFP.Vpr labeled HIV-1 virions with (i) the parental 4E10, (ii) the unmodified ΔLoop Fab or (iii) ΔLoop-Fus4 Fab, and quantified antibody signal. Owing to the linear nature of the STED phenomenon, the number of photons recorded by the detector is directly proportional to the number of bound Fabs (Figure 4A), which permits the estimation of the antibody binding affinity. Consistent with the neutralization data (Figures 1E and 3), almost no binding to native Env was observed when employing ΔLoop Fab. In contrast, the chemically modified ΔLoop-Fus4 antibody showed a binding profile remarkably similar to the parental 4E10 Fab (Figure 4A).
Figure 4.
Fus4 conjugation results in efficient binding to native HIV-1 Env
(A) Top: Schematic representation of the correlation between fluorescence intensity signal (pink-antihuman- STAR RED) and number of Fabs bound to JRCSF-pseudotyped virions (green- Vpr.EGFP) measured by STED microscopy. Representative micrographs with no (top), low (middle) or high number (bottom) of bound antibodies are shown. Right panel: Representative micrographs and quantification of the fluorescence intensity of the bound Fab signal in individual virions from two independent experiments. Virions incubated only with the antihuman STAR RED secondary antibody were added as a negative control. Lines represents the mean value and whiskers the 5th and 95th percentiles. Scale bars in the micrographs are 100 nm.
(B) Binding comparison of Fabs 4E10, ΔLoop and ΔLoop-Fus4 to native Env trimers on HIV-1 virions. Formation of Env-Fab complexes were resolved by BN-PAGE Western blot. The schematic representation depicts the gp41 (blue), gp120 (red) and Fab (green) proteins.
These observations were confirmed using blue native polyacrylamide gel electrophoresis (BN-PAGE) gel mobility shift assays (Leaman and Zwick, 2013) to measure binding to virion-associated Env trimers (Figure 4B). As expected, the observed increase in neutralization with ΔLoop-Fus4 Fab matched with increased binding to virion-associated Env trimers, as there is a similar upward gel mobility shift of the Env spike band upon incubation of virions with ΔLoop-Fus4 and parental 4E10 Fab. The inability of ΔLoop to neutralize the virus is related to a meager capacity of this variant to bind to the Env trimer, as seen by the negligible shifts in the BN-PAGE bands (Figure 4B). Overall, these data confirm that Fus4 replaces the biological function of the hydrophobic residues at the CDRH3 apex of 4E10 and allows binding to native MPER helices, which results in a stable Fab-Env complex and virus neutralization.
Trimming-grafting the CDRH3 loop apex ameliorates the autoreactivity profile of 4E10
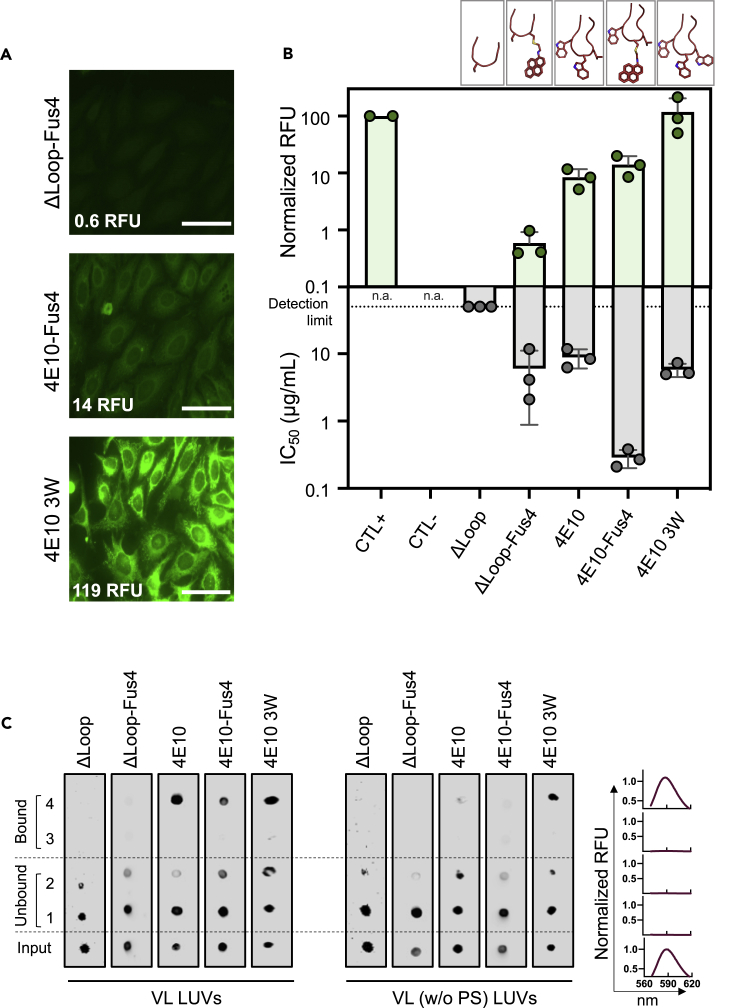
Next, we tested the autoreactivity profile of ΔLoop-Fus4 Fab by measuring binding to HEp-2 epithelial cells (Figures 5A and 5B). Notably, cells treated with the modified ΔLoop-Fus4 Fab showed remarkably low fluorescence intensity levels, resembling the autoreactivity profile of the unmodified ΔLoop variant (Figure 1D). To confirm that concentrating interfacial hydrophobicity at a single position does not translate into enhanced autoreactivity, we replaced W100b of parental 4E10 Fab with Fus4, also by chemical modification, to generate the – W-G-Fus4-L – tip. Consistent with the results obtained for ΔLoop, the presence of Fus4 did not significantly increase the interaction of modified 4E10 with HEp-2 cells with respect to the unmodified antibody. A similar binding profile was obtained for both Fab 4E10 and Fab 4E10-Fus4, in spite of the latter having two additional hydrophobic/aromatic fused rings at the CDRH3 (Figures 5A, 5B and 1D). However, when the interfacial hydrophobicity was distributed through the CDRH3 instead of being concentrated at the H.W100b position, the resulting Fab (4E10-3W) displayed a stronger staining of HEp-2 cells than that observed by the 4E10 Fab, indicating a greater autoreactivity (Figures 5A, 5B and 1D). In this case, the two additional fused rings corresponded to an additional Trp residue at the tip of the CDRH3 loop at the position of the hydrophobic residue Leu (– W-G-W-W – tip).
Figure 5.
Fus4 engraftment onto ΔLoop's CDRH3 does not rescue autoreactivity
(A) Immunofluorescence staining against HEp-2 epithelial cells. Normalized RFU values are indicated. Scale bars in the micrographs are 50 μm.
(B) Quantitative comparison of HEp-2 cells staining and neutralization potency (IC50 neutralization values against JRCSF-pseudotyped virions). Mean ± SD values obtained from three independent experiments are plotted.
(C) Spontaneous water-membrane partitioning measured in vesicle flotation assays. VL composition is compared with VL (-PS) devoid of anionic phospholipid. See also Figures S1 and S2; Table S1.
Notably, this set of CDRH3 mutants further confirmed the apparent lack of correlation between autoreactivity and antibody antiviral (neutralization) activity (Figure 5B). On the one hand, 4E10-Fus4 displayed a 30-fold increase in neutralization potency in comparison to the parental 4E10 without a significant increment on its autoreactivity levels. On the other hand, the potency of the 4E10-3W Fab did not appreciably improve, whereas its autoreactivity levels significantly increased. Thermostability assays demonstrated that the modification of the sequence of the CDRH3 loop did not alter the melting and aggregation temperature of the Fabs, discarding that defects in stability could influence their autoreactivity profile (Figure S2).
The results of Fab binding to VL membranes were also consistent with the above observations (Figure 5C). Following the same trend observed for ΔLoop, the modified ΔLoop-Fus4 Fab did not associate spontaneously with vesicles made of the VL mixture. Similarly, modification of the parental antibody with Fus4 did not change its membrane partitioning behavior compared to that of the parental 4E10 Fab. In both cases, the ability to interact with lipid membranes was dependent on the presence of the anionic phospholipid phosphatidylserine (Figure 5C). In contrast, the 4E10 3W Fab bound to VL membranes also in the absence of PS, highlighting the influence of the hydrophobic component of this Fab in driving membrane binding, and consequently in its autoreactive profile. Overall, the data gathered in this section suggest that strategies to narrow the hydrophobic distribution within the CDRH3 loop could be employed to improve the properties of antibodies directed against membrane embedded epitopes (Figure 6).
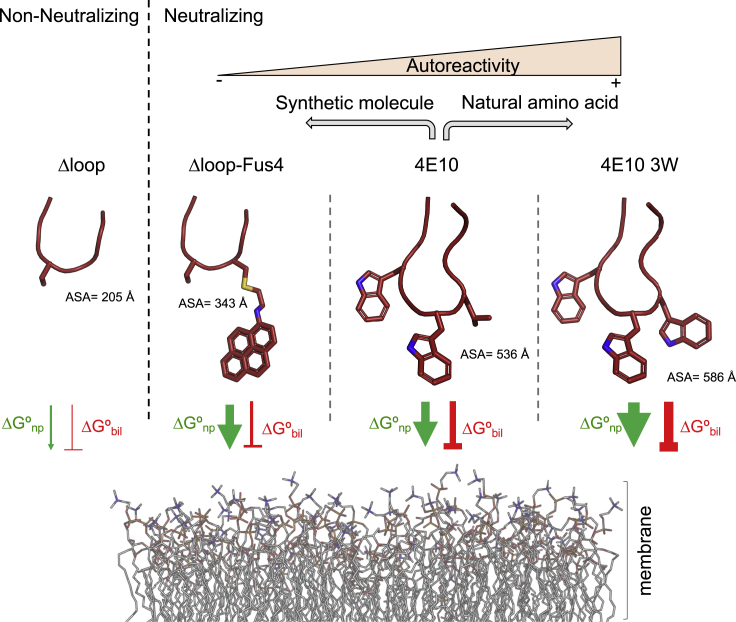
Figure 6.
Model for Fus4 effects on CDRH3 function
A silhouette model of the CDRH3 apex of antibodies ΔLoop, ΔLoop-Fus4, 4E10 and 4E10 3W was employed to illustrate the composition of each loop. The propensity of interacting with lipid membranes was represented as two opposing forces, a favorable energy driven by nonpolar interactions (ΔGnp, green arrow), and an unfavorable energy caused by the bilayer effect (ΔGbil, red line). The accessible surface area (ASA) of the exposed residues was calculated using the PDBePISA server (Krissinel and Henrick, 2007). The calculations were carried out with PDB: 5CIN (for ΔLoop, chain H), and 4XBG (for 4E10, chain A). Models of ΔLoop-Fus4 and 4E10 3W were generated from 5CIN and 4XBG, respectively and ASA calculated accordingly in the PDBePISA server. The autoreactivity outcome of the two strategies, i.e. the use of natural amino acids or synthetic molecules employed to increase the biological properties of 4E10 is shown.
Discussion
Antibody polyreactivity generally correlates with poor in vivo half-life of antibodies (Rudicell et al., 2014; Sievers et al., 2015), requiring their modification by protein engineering to improve their biophysical properties and pharmacokinetic profiles. However, the optimization of sequences aiming to reduce polyreactivity may as well abolish biological activity. Several HIV antibodies targeting the MPER helix of Env harbor a hydrophobic patch of residues at the apex of their CDRH3 loop, which must interact with the viral membrane in order to reach the MPER moiety. Those hydrophobic residues, that are critical for the neutralizing activity of the antibody, also contribute to their unspecific binding to membranes.
In this study, we focused on the HIV-1 antibody 4E10, an anti-MPER antibody that shows some degree of polyreactivity. We aimed at developing a strategy to minimize its polyreactivity, while preserving antibody potency and breadth. We used site-selective chemical modification to concentrate in a single compound (Fus4) the functional equivalent of the hydrophobicity/aromaticity of the four residues at the apex of the CDRH3 – WGWL – of 4E10. This strategy yielded a 4E10 variant with an optimized balance of hydrophobicity in the CDRH3 that evaded unspecific lipid binding while facilitating its functional insertion into the viral membrane. This result agrees with previous studies in which it was postulated that the formation of an induced lipid binding site upon epitope binding accounted for the low polyreactivity of a MPER antibody (Krebs et al., 2019; Zhang et al., 2019). In the following, we discuss possible mechanisms to explain the outcome of this approach.
Insertion of the apex of the 4E10 CDRH3 loop into the viral membrane depends on favorable nonpolar interactions with the water:lipid interface (ΔGnp) but also on the bilayer effects (ΔGbilayer) that usually oppose such insertion. These effects comprise the changes in conformation and reduced mobility of the inserted peptide chain, and the deformation energy of the lipid bilayer (White and Wimley, 1999) (Figure 6). It should be noted that the latter will be of special relevance during insertion of the apex of the loop due to (i) the high degree of flexibility observed in solution for this element in the apo form of the antibody (Rujas et al., 2015) and (ii) the fact that a portion of the non-H-bonded antibody chain must penetrate the membrane interface (White and Wimley, 1999). In the specific case of 4E10, the negative bilayer effect could be partially overcome by the Trp residues at the apex, since their side chains favorably partition into the interfacial region of lipid bilayers (Wimley and White, 1996; Yau et al., 1998) presumably due to their quadrupole moment and planarity (Yau et al., 1998). In this context, we hypothesized that Fus4 concentrates and intensifies those favorable molecular properties in a single moiety and at a single position, thus favoring penetration of the antibody loop in the membrane. In addition, we believe that the bilayer effect would be less intense when employing a CDRH3 loop of reduced size, like in ΔLoop, reducing the entropy loss upon insertion. Hence, these factors could explain the recovery of the biological function after chemical modification of this antibody variant.
Notably, recovery of the biological function of 4E10 took place without a significant development of off-target interactions with bare membranes or with HEp-2 cells. As indicated previously, building favorable interactions between Trp residues and the lipid bilayer interface is expected to contribute favorably to the insertion of the parental 4E10 CDRH3. However, distributing aromaticity along the CDRH3 loop across multiple Trp residues also enlarges the hydrophobic surface of the antibody exposed to the solvent (accessible surface area [ASA] values in Figure 6). We speculate that increasing the surface of this patch beyond a certain level results in the promotion of nonspecific interactions with the membrane, whereas the reduction of the hydrophobic surface accessible to the solvent as shown for ΔLoop-Fus4 is expected to mitigate off-target non-polar interactions of the CDRH3 loop (Figure 6). Thus, aromaticity accretion at a single position of the loop appears to ensure stable Fab association with Env on virions.
Anti-MPER antibodies are important HIV therapeutic candidates due to their extraordinary breadth. However, their clinical use might be limited by their potency (Manrique et al., 2007; Mendoza et al., 2018). Consistent with the importance of antibody potency and breadth, the recent clinical trials HVTN 704/HPTN 085 with the bnAb VRC01 established an IC80 threshold of 1 μg/mL to achieve in vivo protection against an HIV strain (Corey et al., 2021). Hence, the near pan-neutralization breadth of MPER antibodies warrants engineering efforts to improve their potency. In this regard, we have previously shown that site-selective chemical modification of the anti-MPER bnAb 10E8 with aromatic residues dramatically increased its capacity to block viral infection (Rujas et al., 2020). Similarly, here we showed that aromatic grafting in the parental 4E10 resulted in 30-fold improvement of the neutralization potency of this antibody. However, it should be noted that Fus4 could induce anti-drug antibodies in vivo, decreasing its effectiveness and hence the potential impact of Fus4 on antibody immunogenicity needs to be studied. In addition, the need of a conjugation step increases production complexity likely affecting the yield of the final product. These are important aspects that should be considered if this approach were to be followed in developing a clinical product, over more conventional methods previously employed (Kwon et al., 2016).
Finally, several antibodies with a more favorable biological profile than 4E10 have been described, lowering the interest of this antibody as a clinical candidate. However, some of those share the same germline V/D-region genes of 4E10 (Krebs et al., 2019; Zhang et al., 2019). Remarkably, these Abs engage MPER following a common molecular mechanism of recognition, yet they present diverse polyreactivity profiles and some possess shorter CDRH3s. Hence, it will be interesting to explore the potency-enhancement capacity of Fus4 conjugation to bnAbs of the 4E10 class like PGZL1 that has a shorter CDRH3 and reduced polyreactivity compared with 4E10 (Krebs et al., 2019; Zhang et al., 2019), or with other antibodies that target membrane proximal epitopes, to explore if this approach can be applied to more clinically relevant antibodies.
Overall, our study suggests that the CDRH3 of 4E10 might act as a Velcro-like tightening system to form a stable Env-complex incompatible with fusion progression and puts forward the possibility that chemical engineering of CDRH3 hydrophobicity could be used as a general strategy to improve recognition of epitopes proximal to the membrane. In addition, these data provide an alternative strategy to increase hydrophobicity without generating unspecific lipid interactions by concentrating interfacial hydrophobicity at a single position.
Limitations of the study
In this study, a single HIV antibody, i.e., 4E10, has been selected to validate the use of aromatic grafting as a method to preserve/improve binding to membrane embedded epitopes without generating off-target effects. However, we acknowledge that the clinical use of 4E10 is limited due to the emergence of more potent antibodies, and that applying this strategy to a larger set of antibodies will be needed to determine the applicability of this method.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Donkey anti-Human IgG Abberior STAR RED | Abberior | CTRED-1054; RRID:AB_2877172 |
| Goat anti-Human IgG (Fab specific) | Sigma | I5260; RRID:AB_260206 |
| Rabbit anti-human IgG-HRP | Santa Cruz Biotechnology | sc-2769; RRID:AB_656966 |
| Mouse anti-goat IgG-HRP | Santa Cruz Biotechnology | sc-2354; RRID:AB_628490 |
| Bacterial and Virus Strains | ||
| E. coli T7Shuffle | New England Biolabs | C3026J |
| E. Coli DH5α competent cell | Invitrogen | 18265017 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2-Iodo-N-(pyren-1-yl) acetamide (Fus4) | Life Technologies | P29 |
| gp41 MPER peptide | ProteoGenix | N/A |
| 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) | Avanti Polar Lipids | 42773 |
| 1-palmitoyl-2-oleoylphosphatidylethanolamine (POPE) | Avanti Polar Lipids | 01991 |
| 1-palmitoyl-2-oleoylphosphatidylserine (POPS) | Avanti Polar Lipids | 840034C |
| N-palmitoylsphingomyelin (SM) | Avanti Polar Lipids | 85615 |
| Cholesterol (Chol) | Avanti Polar Lipids | C8667 |
| DNase I | PanReac AppliChem | A3778,0010 |
| EDTA-free protease inhibitor mixture | Roche | 11873580001 |
| MEM Non-essential Amino Acid Solution | Thermo Scientific | 11140050 |
| Sodium Pyrubate | Thermo Scientific | 11360070 |
| Opti-MEM I Reduced-Serum Medium | Thermo Scientific | 11058021 |
| KOD-Plus mutagenesis kit | Toyobo | SMK-101 |
| Bright-Glo luciferase reagent | Promega | E2610 |
| PEI, MW 25 kDa | Polysciences | 23966-1 |
| DMEM Growth Medium | Life Technologies | 10313-039 |
| Fetal Bovine Serum | Life Technologies | 10437-028 |
| NuPAGE 3-8% Tris-acetate Gels | Life Technologies | EA03755BOX |
| Tris-glycine Native Sample Buffer | Life Technologies | LC2673 |
| Coomassie Brilliant Blue G250 | Sigma | 27815 |
| n-Dodecyl-beta-Maltoside (DDM) | Pierce | 89903 |
| ECL Plus Western Blotting Substrate | Pierce | 32132 |
| Sinapinic acid [4-Hydroxy-3,5-dimethoxycinnamic acid] MALDI matrix | Alfa Aesar | A15676 |
| Trifluoroacetic acid | Thermo scientific | 28904 |
| Water, Optima LC/MS grade | Fischer chemical | W6-1 |
| Acetonitrile, Optima LC/MS grade | Fischer chemical | A955-212 |
| Protein Calibration Standard 1 mixture | Bruker Daltonics | 206355 |
| Poly-L-lysine solution | Sigma-Aldrich | P8920 |
| Bovine Serum Albumin (fatty acid free) | Sigma-Aldrich | A7030 |
| PBS1x without calcium and magnesium | Thermo Fisher | Cat#12037539 |
| PenStrep | Gibco | Cat#15140-122 |
| Critical Commercial Assays | ||
| Kallestad HEp-2 Kits | BioRad | 30471 |
| Deposited Data | ||
| 4E10-Fus4 | Protein Data Bank | 7EKB |
| Δloop-Fus4 | Protein Data Bank | 7EKK |
| Experimental Models: Cell Lines | ||
| Human: HEK293T Cells | ATCC | CRL-3216 |
| Human: TZM-bl Cells | NIH AIDS Reagent Program | 8129 |
| Recombinant DNA | ||
| pCOLADuet-1 expression plasmid | Sigma-Aldrich | 71406 |
| 4E10 Fab pCOLADuet-1 wild-type and mutants | This paper | N/A |
| pWXLP-GFP | Patricia Villacé, CSIC | N/A |
| pCMV8.91 | Patricia Villacé, CSIC | N/A |
| PVO, clone 4 (SVPB11) Expression plasmid | NIH AIDS Reagent Program | 11022 |
| JRCSF Expression plasmid | Jamie Scott, SFU | N/A |
| pSG3ΔEnv | NIH AIDS Reagent Program | 11051 |
| ADA.CM Expression plasmid | Leaman and Zwick, 2013 | N/A |
| 92RW020.5 Expression plasmid | NIH AIDS Reagent Program | 3097 |
| 94UG103 Expression plasmid | D. Burton, Scripps (Simek et al., 2009) | N/A |
| 92BR020 Expression plasmid | NIH AIDS Reagent Program | 1780 |
| 92TH021 Expression plasmid | D. Burton, Scripps (Simek et al., 2009) | N/A |
| IAVI C22 Expression plasmid | D. Burton, Scripps (Simek et al., 2009) | N/A |
| BG505 Expression plasmid | NIH AIDS Reagent Program | 11518 |
| P16055 Expression plasmid | NIH AIDS Reagent Program | N/A |
| pEGFP.Vpr | NIH AIDS Reagent Program | 11386 |
| Software and Algorithms | ||
| GraphPad Prism 8 | GraphPad | N/A |
| Autoflex III | Bruker Daltonics | N/A |
| Autoflex III | Bruker Daltonics | N/A |
| Pymol Molecular Graphics System | Schrödinger | N/A |
| Imspector software | Abberior Instruments | N/A |
| STED analysis program | Galiani et al., 2016 | https://doi.org/10.5281/zenodo.1465920. |
| OriginPro 2019b (9.6.5.169) | OriginLab Corporation | N/A |
| biorender | biorender.com | https://biorender.com/ |
| Microcal Origin 7.0 | OriginLab | N/A |
| UNit analysis software | Unchained Labs | N/A |
| Coot | Emsley et al., 2010 | N/A |
| PDBePISA | Krissinel and Henrick, 2007 | N/A |
| UCSF Chimera | Pettersen et al., 2004 | N/A |
| Other | ||
| ZipTip® C4 micro-columns | Millipore | ZTC04S096 |
| Ground Steel massive 384 MALDI target plate | Bruker Daltonics | 8074115 |
| Nickel-nitrilotriacetic acid (Ni-NTA) affinity column | GE-Healthcare | 10431065 |
| MonoS ion exchange chromatography (IEC) column | GE-Healthcare | 17516801 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jose Luis Nieva (joseluis.nieva@ehu.es).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement
Experimental model and subject details
Escherichia coli T7-shuffle strain was grown in Luria Broth medium following the specifications of the supplier.
HEK293T and TZM-bl cells were cultured in DMEM (Dulbecco's Modified Eagle Medium), supplemented with 1 mM sodium pyruvate, 1 mM MEM Non-Essential Amino Acids Solution and 10% FBS, in a humidified incubator at +37°C, supplied with 5% CO2. For cell seeding, cells were washed with phosphate buffer saline (PBS) and then detached with trypsin/EDTA solution (Thermofisher).
Method details
Production and site-selective chemical modification of Fabs
Antibody Fab sequences were cloned in the plasmid pColaDuet and expressed in Escherichia coli T7-shuffle strain. Recombinant expression was induced at 18°C overnight with 0.4 mM isopropyl-D-thiogalactopyranoside when the culture reached an optical density of 0.8. Cells were harvested and centrifuged at 8,000 × g, after which they were resuspended in a buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 35 mM imidazole, DNase (Sigma-Aldrich, St. Louis, MO) and an EDTA-free protease inhibitor mixture (Roche, Madrid, Spain). Cell lysis was performed using an Avestin Emulsiflex C5 homogenizer. Cell debris was removed by centrifugation, and the supernatant loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) affinity column (GE Healthcare). Elution was performed with 500 mM imidazole, and the fractions containing the His-tagged proteins were pooled, concentrated and dialyzed against 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 1 mM DTT, and 0.3 mM EDTA in the presence of purified protease Tobacco etch virus (Kawai et al., 2011). Fabs were separated from the cleaved peptides containing the His6x tag by an additional step in a Ni-nitrilotriacetic column. The flow-through fraction containing the antibody was dialyzed overnight at 4°C against sodium acetate (pH 5.6) supplemented with 10% glycerol and subsequently loaded onto a MonoS ion-exchange chromatography (IEC) column (GE Healthcare). Elution was carried out with a gradient of potassium chloride and the fractions containing the purified Fab concentrated and dialyzed against a buffer containing 10 mM sodium phosphate (pH 7.5), 150 mM NaCl, and 10% glycerol.
Site-selective chemical conjugation was carried out as previously described (Rujas et al., 2020). In brief, a cysteine-substituted Fab derivative was first generated by site-directed mutagenesis using the KOD-Plus mutagenesis kit (Toyobo, Osaka, Japan). Mutation H.S100C and W100bC was generated in the case of ΔLoop-Fus4 and 4E10-Fus4, respectively. Next, the mutated Fabs were conjugated to the sulfhydryl-specific iodoacetamide derivative of the compound Fus4 upon incubation of the Fabs at 37°C overnight with a 10× excess of the compound. Conjugation was monitored by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry.
Isothermal titration calorimetry (ITC)
ITC experiments were performed with a VP-ITC microcalorimeter (MicroCal, Northampton, MA) at 20°C. Fabs of 4E10 or ΔLoop each at 3 μM previously dialyzed against PBS buffer (10 mM sodium phosphate (pH 7.5), 150 mM NaCl) supplemented with 10% (v/v) glycerol were titrated with 40 μM peptide resuspended in the same buffer. Peptide dilution heat was subtracted for data analysis. The binding isotherms were fitted to a one-site binding model using the program ORIGIN 7.0 (MicroCal, Northampton, MA). The fitting procedure yielded the stoichiometry (n) and the binding constant (KD) of the binding reaction.
Thermostability assay
The melting temperature (Tm) and aggregation temperature (Tagg) of the different Fabs was determined using a UNit system (Unchained Labs). Samples were prepared at 1 mg/mL in 10 mM sodium phosphate (pH 7.5), 150 mM NaCl, and 10% (v/v) glycerol and subjected to a thermal ramp from 25 to 95°C with 1°C increments. Tm values were calculated from the barycentric mean fluorescence. On the other hand, Tagg values were determined as the temperature at which 50% increase in the static light scattering at a 266 nm wavelength relative to baseline was observed. The average and the standard error of two measurements were calculated using the UNit analysis software.
Mass spectrometry analysis
Prior to the measurement, conjugated antibodies were desalted using ZipTip® C4 micro columns (Millipore) (2 μL sample) with 0.5μL SA buffer (sinapinic acid, 10 mg/mL in [70:30] Acetonitrile:Trifluoroacetic acid 0.1%), and spotted onto a Ground Steel massive 384 target plate (Bruker Daltonics). Mass determination was performed by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) analysis in an Autoflex III (Bruker Daltonics). Mass calibration was performed externally with a Protein Calibration Standard 1 (Bruker Daltonics) mixture in the same mass range as the samples. Data acquisition, peak peaking and subsequent spectra analysis were performed using flexAnalysis 3.0 software (Bruker Daltonics).
Polyreactivity assays
HIV-1 negative human epithelial HEp-2 (Kallestad ANA/HEp-2) were incubated with 25 μL from a Fab stock solution diluted to 50 μg/mL. Anti-Human IgG (Fab specific)−FITC antibody produced in goat was used as the secondary antibody. Images were acquired on a Leica TCS SP5II microscope (Leica Microsystems GmbH, Wetzlar, Germany). Cells were imaged through a ×63 water-immersion objective (NA = 1.2) and 1024 × 1024 pixel images were acquired at 200 Hz per scanning line. Images obtained under the same gain/detector conditions were subsequently analyzed using the FIJI distribution of ImageJ (Schindelin et al., 2012) as follows: First, pixel intensities within several background regions were measured to estimate the average value of the background; next, taken such value as threshold, the average value of the pixel intensities above that threshold was measured for each sample, and verified that corresponded to areas covered by cells; intensity values were finally referred to a 0–100% scale, were 0% and 100% relative light unit values correspond respectively to those of the negative and positive ANA controls provided by the manufacturer.
Direct interaction with membranes was evaluated in vesicle flotation experiments as previously described (Rujas et al., 2018). In brief, large unilamellar vesicles were prepared at a concentration of 1.5 mM lipid of the VL mixture DOPC:DOPE:DOPS:SM:Chol (14:16:7:17:46) (Scherer et al., 2010). 100 μL of a sample containing 0.5 mol % Rho-PE-labeled VL vesicles was adjusted to a sucrose concentration of 1.4 M in a final volume of 300 μL, and subsequently overlaid with 400 μL- and 300 μL-layers of 0.8 M and 0.5 M sucrose, respectively. The gradient was centrifuged at 436,000 × g for 3 h at 4°C in a TLA 120.2 rotor (Beckman Coulter). After centrifugation, four 250 μL-fractions were collected. The presence of Fabs in the different fractions and in the original sample (input) was revealed by Western Blot analysis after Tris-Tricine SDS-PAGE separation. The presence of liposomes in the different fractions was determined after measuring the fluorescence emission of the Rho-PE probe (Thermo Fisher Scientific, Waltham, Massachusetts, USA)
Crystallization, data collection and processing, and structure refinement
The 4E10-Fus4:peptide complex at 3 mg mL−1 in PBS buffer supplemented with 10% glycerol was crystallized in a solution composed of 200 mM ammonium sulfate, 100 mM sodium acetate (pH 4.6) and 12% PEG 4,000 by the method of sitting-drop. Similarly, the ΔLoop-Fus4:peptide complex at 3 mg mL−1 in PBS buffer supplemented with 10% glycerol was crystallized in a solution composed of 200 mM ammonium sulfate and PEG 4,000, also using the method of sitting-drop. Suitable crystals of 4E10-Fus4 or ΔLoop-Fus4 in complex with epitope peptide were harvested, briefly incubated in mother liquor supplemented with 20–30% (v:v) glycerol, and transferred to liquid nitrogen for storage until data collection. Diffraction data from single crystals were collected in beamline XALOC at the ALBA synchrotron (Cerdanyola del Valles, Spain) under cryogenic conditions (100 K). Diffraction images were processed with the program MOSFLM and merged and scaled with the program SCALA (Evans, 2006) of the CCP4 suite (Winn et al., 2011). The structure of the complex of epitope peptide with 4E10-Fus4 and ΔLoop-Fus4 were determined by the molecular replacement method using the coordinates of unmodified 4E10 (PDB entry code 4WY7) (Apellaniz et al., 2015) and the coordinates of ΔLoop (5CIN) (Rujas et al., 2015), respectively, with the program PHASER (McCoy et al., 2007). The models were refined with the programs REFMAC5 (Murshudov et al., 1997) and refined manually with COOT (Emsley et al., 2010). Validation was carried out with PROCHECK (Laskowski et al., 1993). Other structure calculations were carried out with CHIMERA (Pettersen et al., 2004). Data collection and structure refinement statistics are given in Table S1.
Virus production and neutralization assays
Antibody potency was evaluated in pseudovirus (PsV)-based cell-entry assays (Louder et al., 2005). HIV-1 PsVs were produced by transfection of human kidney HEK293T cells with the full-length Env clone JRCSF (kindly provided by Jamie K. Scott and Naveed Gulzar, Simon Fraser University, BC, Canada) and the PVO.4 molecular clone (obtained from the AIDS Research and Reference Reagent Program (ARRRP)). Cells were co-transfected with vectors pWXLP-GFP and pCMV8.91, encoding a green fluorescent protein and an Env-deficient HIV-1 genome, respectively (provided by Patricia Villace, CSIC, Madrid, Spain). After 24 h, the medium was replaced with Optimem-Glutamax II (Invitrogen Ltd, Paisley, UK) without serum. Two days after transfection, the PsV particles were harvested, passed through 0.45 μm pore sterile filters (Millex® HV, Millipore NV, Brussels, Belgium) and finally concentrated by ultracentrifugation in a sucrose gradient. HIV entry was determined using CD4+CXCR4+CCR5+ TZM-bl target cells (ARRRP, contributed by J. Kappes). To that end, HIV PsVs were first diluted to 10–20% tissue culture infectious doses in DMEM supplemented with inactive serum, and added to the modified and WT Fabs. Infection levels after 72 hours were inferred from the number of GFP-positive cells as determined by flow cytometry using a BD FACSCalibur Flow Cytometer (Becton Dickinson Immunocytometry Systems, Mountain View, CA).
The ability of Fab to inhibit infection by a diversity of HIV isolates was tested using a previously described neutralization assay and panel of PsVs (Rujas et al., 2018). Briefly, HIV-1 PsVs, were produced by co-transfection of Env-complementation plasmid and the HIV-1 backbone plasmid pSG3ΔEnv in HEK293T cells. Fabs, diluted in cell culture media starting at 50 μg/mL and proceeding in a 5-fold dilution series, were added to PsVs and incubated for 1h at 37°C. Fab/virion mixtures were then added to TZM-bl target cells and further incubated for 72 h at 37°C. Cells were lysed and luciferase intensity was read using Bright-Glo luciferase reagent (Promega) and a Synergy H1 plate reader (BioTek).
Binding to native Env by super-resolution fluorescence STED microscopy of single virions
Concentrated JRCSF pseudotyped virus particles containing Vpr.EGFP were adhered to poly-L-lysine (Sigma) coated glass coverslips for 15 min. Then, coverslips were blocked using 2% fatty acid free bovine serum albumin (Sigma)/PBS for 15 min. Fabs (25 ng/μL) were incubated for 1 h in blocking buffer at a 25 ng/μL concentration and revealed upon incubation with anti-human Abberior STAR RED (Abberior GmbH) conjugated Ab. The sample was washed and mounted in PBS, followed by STED imaging. All steps were carried out at RT.
Imaging was performed on a STED microscope based on a modified Abberior Instrument RESOLFT QUAD-P super-resolution microscope (Abberior Instruments GmbH). The sample was excited and the emission collected using a 100×/1.40 NA UPlanSApo oil immersion objective (Olympus Industrial, Southend-on-Sea, UK). The fluorescence signal was descanned, passed through an adjustable pinhole (Thorlabs Limited, Ely, UK) and detected by single photon counting avalanche photo diodes (SPCM-AQRH-13, Excelitas Technologies) with appropriate fluorescence filters (AHF Analysentechnik). All acquisition operations were controlled by Imspector software (Abberior Instruments). Resolution was typically around 40 nm. Emitted photons were recorded line by line in STED microscopy mode, and Vpr.eGFP was next imaged in confocal mode to determine the location of HIV-1 virions.
Image analysis was performed using Python scripting language and custom written functions (Carravilla et al., 2019; Galiani et al., 2016). Individual virions were identified from the Vpr.eGFP channel using an intensity maximum finding algorithm on a Gaussian smoothed image (σ = 2.0). A circular region (diameter, 20 pixels; 400 nm) was then superimposed on each detected location, and the STAR RED signal was quantified at said region and compared to a random region within 90-pixels constrained not to present Vpr.EGFP signal. To permit comparison between independent experiments, the STAR RED emission signal was normalized to that of the median intensity value of the 4E10 sample.
Blue native (BN)-PAGE Western Blot
BN-PAGE gel mobility shift assays were performed using virions produced by transfection of the infectious molecular clone plasmid pLAI displaying the Env variant ADA.CM (Leaman and Zwick, 2013). Virions and a dilution series of Fab were incubated at RT for 30 min. The virion/Ab mixture was solubilized in 1% n-Dodecyl β-D-maltoside (DDM; Sigma) for 20 min on ice, and then run on a 3–8% gradient Tris-Acetate NuPAGE gel (ThermoFisher) in Tris-Glycine Native Sample Buffer (ThermoFisher), supplemented with 0.25% Coomassie G-250. Gels were run for 3 h at 150 V in Tris-Glycine Native Running Buffer (ThermoFisher) + 0.002% Coomassie G-250. Proteins were transferred onto PDVF membrane and Western blotted using a cocktail of primary antibodies to gp120 (F105, 2G12 and HGN194, 2 μg/mL each) and gp41 (10E8, 2F5, and 7B2, 1 μg/mL each) followed by a goat anti-human-Fcγ-HRP secondary antibody (Jackson). The blot was developed using ECL Plus Substrate (Pierce) and a ChemiDoc XRS + Imaging System (Bio-Rad).
Acknowledgments
This study was supported by the following Grants: European Commission (790012 SI H2020-MSCA-IF-2017) (E.R.); US NIAID, NIH grant R01 AI143563 (M.B.Z.); James B. Pendleton Charitable Trust (M.B.Z.); JSPS grant 20H03228 (J.M.M.C.); Spanish MCIU (RTI2018-095624-B-C21; MCIU/AEI/FEDER, UE) (J.L.N.), Basque Government (IT1196-19) (J.L.N.). C.E. acknowledges funding from Medical Research Council (grant number MC_UU_12010/unit programs G0902418 and MC_UU_12025), Wolfson Foundation, Deutsche Forschungsgemeinschaft (Excellence Cluster Balance of the Microverse, Collaborative Research Center 1278 Polytarget), Leibniz Association (Leibniz Campus Infectooptics), Wellcome Institutional Strategic Support Fund, Oxford internal funds (EPA Cephalosporin Fund and John Fell Fund), and support from the Micron Oxford Advanced Bioimaging Unit (Wellcome Trust funding 107457/Z/15/Z). This work was also supported by the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED (JP21am0101091). S.I. received a pre-doctoral fellowship from the Basque Government. P.C. would like to acknowledge the University of the Basque Country (DOCREC18/01), the Basque Government (POS_2018_1_0066) and the European Commission (H2020-MSCA-IF-2019-ST project 892232 FILM-HIV) for funding his position. This research was also supported by the CIFAR Azrieli Global Scholar program (J-P.J.), the Ontario Early Researcher Awards program (J-P.J.), and the Canada Research Chairs program (J-P.J.). Part of the biophysical data presented in this manuscript were collected at the Hospital for Sick Children Structural & Biophysical Core facility supported by the Canada Foundation for Innovation and Ontario Research Fund.
Author contributions
E.R., J.M.M.C., and J.L.N. designed research; E.R., D.P.L., S.I., P.C., M.G.-P., E.L., I.M., R. S.-E., L.Z., H.C., and I.I. performed research; E.R., D.P.L., S.I., P.C., M.G.-P., E.L., I.M., R. S.-E., L.Z., H.C., I.I., F.E., J-P.L., C.E., M.B.Z., J.M.M.C., and J.L.N. analyzed data; and E.R., J.M.M.C., and J.L.N. wrote the paper with input from all authors.
Declaration of interests
A patent application covering several aspects of this work has been filled by Kyushu University/UPV-EHU/CSIC/FBB, listing E.R., R.S.-E., J.L.N., and J.M.M.C. as inventors. All other authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102987.
Contributor Information
Edurne Rujas, Email: edurne.rujas@ehu.eus.
Jose M.M. Caaveiro, Email: jose@phar.kyushu-u.ac.jp.
José L. Nieva, Email: joseluis.nieva@ehu.es.
Supplemental information
Data and code availability
The authors declare that all relevant data are available within the article and its Supplemental information files or from the corresponding author upon reasonable request. The crystal structures of 4E10-Fus4 and Δloop-Fus4 (Table S1) are available from the Protein Data Bank under accession codes 7EKB and 7EKK, respectively. This study did not generate new code.
References
- Alam S.M., McAdams M., Boren D., Rak M., Scearce R.M., Gao F., Camacho Z.T., Gewirth D., Kelsoe G., Chen P. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.M., Morelli M., Dennison S.M., Liao H.X., Zhang R., Xia S.M., Rits-Volloch S., Sun L., Harrison S.C., Haynes B.F. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U S A. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alving C.R. 4E10 and 2F5 monoclonal antibodies: binding specificities to phospholipids, tolerance, and clinical safety issues. Aids. 2008;22:649–651. doi: 10.1097/QAD.0b013e3282f51922. [DOI] [PubMed] [Google Scholar]
- Apellaniz B., Rujas E., Serrano S., Morante K., Tsumoto K., Caaveiro J.M., Jimenez M.A., Nieva J.L. The atomic structure of the HIV-1 gp41 transmembrane domain and its connection to the immunogenic membrane-proximal external region. J. Biol. Chem. 2015;290:12999–13015. doi: 10.1074/jbc.M115.644351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan M., Rudicell R.S., Louder M., McKee K., O'Dell S., Stewart-Jones G., Wang K., Xu L., Chen X., Choe M. Bispecific antibodies targeting different epitopes on the HIV-1 envelope exhibit broad and potent neutralization. J. Virol. 2015;89:12501–12512. doi: 10.1128/JVI.02097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carravilla P., Chojnacki J., Rujas E., Insausti S., Largo E., Waithe D., Apellaniz B., Sicard T., Julien J.P., Eggeling C. Molecular recognition of the native HIV-1 MPER revealed by STED microscopy of single virions. Nat. Commun. 2019;10:78. doi: 10.1038/s41467-018-07962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carravilla P., Darre L., Oar-Arteta I.R., Vesga A.G., Rujas E., de Las Heras-Martinez G., Domene C., Nieva J.L., Requejo-Isidro J. The bilayer collective properties govern the interaction of an HIV-1 antibody with the viral membrane. Biophys. J. 2020;118:44–56. doi: 10.1016/j.bpj.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L., Gilbert P.B., Juraska M., Montefiori D.C., Morris L., Karuna S.T., Edupuganti S., Mgodi N.M., deCamp A.C., Rudnicki E. Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N. Engl. J. Med. 2021;384:1003–1014. doi: 10.1056/NEJMoa2031738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellavance A., Andrade L.E.C. Detection of autoantibodies by indirect immunofluorescence cytochemistry on Hep-2 cells. Methods Mol. Biol. 2019;1901:19–46. doi: 10.1007/978-1-4939-8949-2_3. [DOI] [PubMed] [Google Scholar]
- Dennison S.M., Stewart S.M., Stempel K.C., Liao H.X., Haynes B.F., Alam S.M. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. J. Virol. 2009;83:10211–10223. doi: 10.1128/JVI.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. Sect D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Ferrantelli F., Hofmann-Lehmann R., Rasmussen R.A., Wang T., Xu W., Li P.L., Montefiori D.C., Cavacini L.A., Katinger H., Stiegler G. Post-exposure prophylaxis with human monoclonal antibodies prevented SHIV89.6P infection or disease in neonatal macaques. Aids. 2003;17:301–309. doi: 10.1097/00002030-200302140-00003. [DOI] [PubMed] [Google Scholar]
- Galiani S., Waithe D., Reglinski K., Cruz-Zaragoza L.D., Garcia E., Clausen M.P., Schliebs W., Erdmann R., Eggeling C. Super-resolution microscopy reveals compartmentalization of peroxisomal membrane proteins. J. Biol. Chem. 2016;291:16948–16962. doi: 10.1074/jbc.M116.734038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Fleming J., St Clair E.W., Katinger H., Stiegler G., Kunert R., Robinson J., Scearce R.M., Plonk K., Staats H.F. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Nicely N.I., Alam S.M. HIV-1 autoreactive antibodies: are they good or bad for HIV-1 prevention? Nat. Struct. Mol. Biol. 2010;17:543–545. doi: 10.1038/nsmb0510-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell A.J., Rakasz E.G., Tehrani D.M., Huber M., Weisgrau K.L., Landucci G., Forthal D.N., Koff W.C., Poignard P., Watkins D.I. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J. Virol. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yu J., Lanzi A., Yao X., Andrews C.D., Tsai L., Gajjar M.R., Sun M., Seaman M.S., Padte N.N. Engineered bispecific antibodies with exquisite HIV-1-Neutralizing activity. Cell. 2016;165:1621–1631. doi: 10.1016/j.cell.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia A., Sarkar A., Stanfield R.L., Wilson I.A. Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity. 2016;44:21–31. doi: 10.1016/j.immuni.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Caaveiro J.M.M., Abe R., Katagiri T., Tsumoto K. Catalytic activity of MsbA reconstituted in nanodisc particles is modulated by remote interactions with the bilayer. FEBS Lett. 2011;585:3533–3537. doi: 10.1016/j.febslet.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Klein F., Mouquet H., Dosenovic P., Scheid J.F., Scharf L., Nussenzweig M.C. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs S.J., Kwon Y.D., Schramm C.A., Law W.H., Donofrio G., Zhou K.H., Gift S., Dussupt V., Georgiev I.S., Schatzle S. Longitudinal analysis reveals early development of three MPER-directed neutralizing antibody lineages from an HIV-1-Infected individual. Immunity. 2019;50:677–691 e613. doi: 10.1016/j.immuni.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kunert R., Wolbank S., Stiegler G., Weik R., Katinger H. Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res. Hum. Retroviruses. 2004;20:755–762. doi: 10.1089/0889222041524571. [DOI] [PubMed] [Google Scholar]
- Kwon Y.D., Georgiev I.S., Ofek G., Zhang B., Asokan M., Bailer R.T., Bao A., Caruso W., Chen X., Choe M. Optimization of the solubility of HIV-1-Neutralizing antibody 10E8 through somatic variation and structure-based design. J. Virol. 2016;90:5899–5914. doi: 10.1128/JVI.03246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Leaman D.P., Zwick M.B. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. Plos Pathog. 2013;9:e1003184. doi: 10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louder M.K., Sambor A., Chertova E., Hunte T., Barrett S., Ojong F., Sanders-Buell E., Zolla-Pazner S., McCutchan F.E., Roser J.D. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology. 2005;339:226–238. doi: 10.1016/j.virol.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Manrique A., Rusert P., Joos B., Fischer M., Kuster H., Leemann C., Niederost B., Weber R., Stiegler G., Katinger H. In vivo and in vitro escape from neutralizing antibodies 2G12, 2F5, and 4E10. J. Virol. 2007;81:8793–8808. doi: 10.1128/JVI.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyas G.R., Beck Z., Karasavvas N., Alving C.R. Lipid binding properties of 4E10, 2F5, and WR304 monoclonal antibodies that neutralize HIV-1. Biochim. Biophys. Acta. 2009;1788:660–665. doi: 10.1016/j.bbamem.2008.11.015. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald S.K., Fleming K.G. Aromatic side chain water-to-lipid transfer free energies show a depth dependence across the membrane normal. J. Am. Chem. Soc. 2016;138:7946–7950. doi: 10.1021/jacs.6b03460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza P., Gruell H., Nogueira L., Pai J.A., Butler A.L., Millard K., Lehmann C., Suarez I., Oliveira T.Y., Lorenzi J.C.C. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature. 2018;561:479–484. doi: 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Pegu A., Yang Z.Y., Boyington J.C., Wu L., Ko S.Y., Schmidt S.D., McKee K., Kong W.P., Shi W., Chen X. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci. Transl Med. 2014;6:243ra288. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto D., Fenwick C., Caillat C., Silacci C., Guseva S., Dehez F., Chipot C., Barbieri S., Minola A., Jarrossay D. Structural basis for broad HIV-1 neutralization by the MPER-specific human broadly neutralizing antibody LN01. Cell Host Microbe. 2019;26:623–637 e628. doi: 10.1016/j.chom.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantalainen K., Berndsen Z.T., Antanasijevic A., Schiffner T., Zhang X., Lee W.H., Torres J.L., Zhang L., Irimia A., Copps J. HIV-1 envelope and MPER antibody structures in lipid assemblies. Cell Rep. 2020;31:107583. doi: 10.1016/j.celrep.2020.107583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudicell R.S., Kwon Y.D., Ko S.Y., Pegu A., Louder M.K., Georgiev I.S., Wu X., Zhu J., Boyington J.C., Chen X. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujas E., Gulzar N., Morante K., Tsumoto K., Scott J.K., Nieva J.L., Caaveiro J.M. Structural and thermodynamic basis of epitope binding by neutralizing and nonneutralizing forms of the anti-HIV-1 antibody 4E10. J. Virol. 2015;89:11975–11989. doi: 10.1128/JVI.01793-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujas E., Insausti S., Garcia-Porras M., Sanchez-Eugenia R., Tsumoto K., Nieva J.L., Caaveiro J.M. Functional contacts between MPER and the anti-HIV-1 broadly neutralizing antibody 4E10 extend into the Core of the membrane. J. Mol. Biol. 2017;429:1213–1226. doi: 10.1016/j.jmb.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujas E., Insausti S., Leaman D.P., Carravilla P., Gonzalez-Resines S., Monceaux V., Sanchez-Eugenia R., Garcia-Porras M., Iloro I., Zhang L. Affinity for the interface underpins potency of antibodies operating in membrane environments. Cell Rep. 2020;32:108037. doi: 10.1016/j.celrep.2020.108037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujas E., Leaman D.P., Insausti S., Ortigosa-Pascual L., Zhang L., Zwick M.B., Nieva J.L. Functional optimization of broadly neutralizing HIV-1 antibody 10E8 by promotion of membrane interactions. J. Virol. 2018;92 doi: 10.1128/JVI.02249-17. e02249–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M., Bailer R.T., Turk E., Lin C.L., Bilska M., Greene K.M., Gao H., Todd C.A., Ozaki D.A., Seaman M.S. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer E.M., Leaman D.P., Zwick M.B., McMichael A.J., Burton D.R. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc. Natl. Acad. Sci. U S A. 2010;107:1529–1534. doi: 10.1073/pnas.0909680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers S.A., Scharf L., West A.P., Jr., Bjorkman P.J. Antibody engineering for increased potency, breadth and half-life. Curr. Opin. HIV AIDS. 2015;10:151–159. doi: 10.1097/COH.0000000000000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek M.D., Rida W., Priddy F.H., Pung P., Carrow E., Laufer D.S., Lehrman J.K., Boaz M., Tarragona-Fiol T., Miiro G. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D., Burton D.R. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 2018;19:1179–1188. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler G., Katinger H. Therapeutic potential of neutralizing antibodies in the treatment of HIV-1 infection. J. Antimicrob. Chemother. 2003;51:757–759. doi: 10.1093/jac/dkg172. [DOI] [PubMed] [Google Scholar]
- Stiegler G., Kunert R., Purtscher M., Wolbank S., Voglauer R., Steindl F., Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Walker L.M., Burton D.R. Passive immunotherapy of viral infections: 'super-antibodies' enter the fray. Nat. Rev. Immunol. 2018;18:297–308. doi: 10.1038/nri.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Williams L.D., Ofek G., Schatzle S., McDaniel J.R., Lu X., Nicely N.I., Wu L., Lougheed C.S., Bradley T., Louder M.K. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci. Immunol. 2017;2:eaa12200. doi: 10.1126/sciimmunol.aal2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W.C., White S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau W.M., Wimley W.C., Gawrisch K., White S.H. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- Zhang L., Irimia A., He L., Landais E., Rantalainen K., Leaman D.P., Vollbrecht T., Stano A., Sands D.I., Kim A.S. An MPER antibody neutralizes HIV-1 using germline features shared among donors. Nat. Commun. 2019;10:5389. doi: 10.1038/s41467-019-12973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all relevant data are available within the article and its Supplemental information files or from the corresponding author upon reasonable request. The crystal structures of 4E10-Fus4 and Δloop-Fus4 (Table S1) are available from the Protein Data Bank under accession codes 7EKB and 7EKK, respectively. This study did not generate new code.