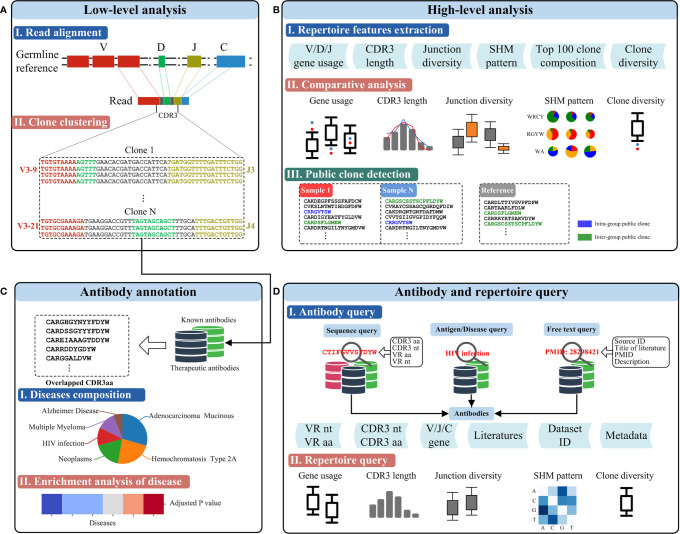
Figure 2.
Functionalities of RAPID. (A) Low-level analysis. Germline genes and the antibody sequence derived from the recombination are shown schematically on top. CDR3s were identified using the RAPID bioinformatics pipeline, and clonalities of antibodies were defined according to sequence similarity and V/J gene segments (bottom). (B) High-level analysis of Rep-seq dataset. Repertoire features were extracted from the Rep-seq datasets (top). The features of the experimental and reference groups were compared and shown (middle). Public clones, if available, were extracted and displayed (bottom). (C) Antibody annotation based on CDR3 aa. Antibodies having the same amino acid CDR3 as known or therapeutic antibodies were extracted and annotated based on their matches in the database (middle). Enrichment of the annotated antibodies were analysed, and a p value was calculated. (D) Antibody and repertoire query function. The top panel shows several antibody queries and the schematics of the result. The bottom panel shows the visualized results of a repertoire query.