Figure 2.
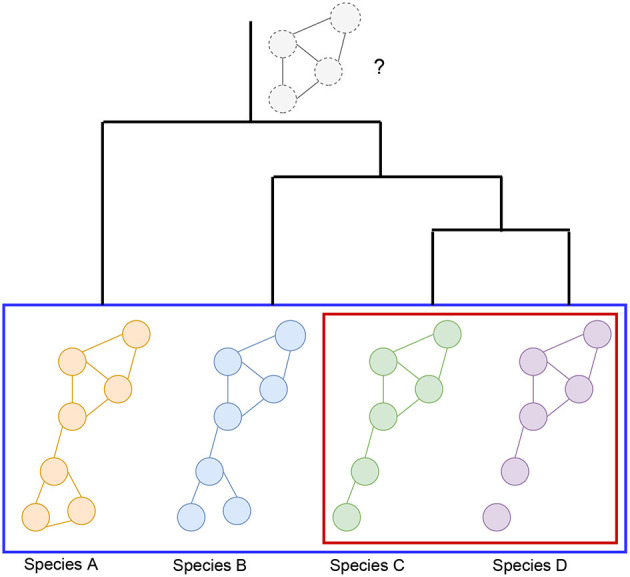
Visualization of the hypothetical changes in a biological network, such as a co-expression network, generated from four different species (species A, B, C, and D). Different comparisons that can be made include the pairwise comparison between the GCNs of two species (red box), or simultaneous comparisons can be made across many species (blue box). These types of comparisons may also be used to infer ancestral GCNs, which may be inferred as portions of the networks that are more conserved across the species being compared. The known information about phylogeny and gene age can also be utilized in order to reconstruct hypothetical ancestral GCNs, such as the one represented at the root of this phylogenetic tree. It is important to note that the ancestral GCN is completely preserved across these species for the sake of this example, but this is unlikely without any evolutionary changes over time. In this alternative scenario, the relationships between the bottom module nodes would not be preserved, suggesting that perhaps whatever this module of genes was responsible for biologically is no longer important for the later diverged species.
