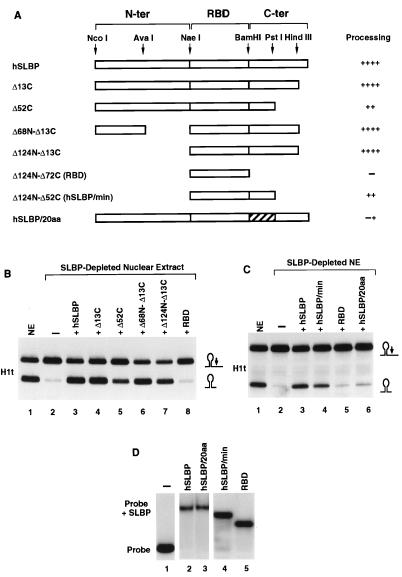
FIG. 4.
Regions of human SLBP required for histone pre-mRNA processing. (A) The restriction map of the human SLBP cDNA with the boundaries of the three regions of the protein corresponding to the N-terminal domain (N-ter), the RNA binding domain (RBD), and the C-terminal domain (C-ter) is outlined at the top. The constructs are named according to the number of amino acids deleted from the N or C terminus. The hSLBP/20aa has the 20 amino acids immediately after the RNA binding domain (196 to 215) changed to the 20 amino acids present at this position in frog XSLBP2, an SLBP which does not function in pre-mRNA processing (43). The ability of the mutant SLBP to restore processing in the SLBP-depleted extract is summarized to the right of each construct. (B) The nuclear extract (lane 1) was depleted of SLBP with anti-SLBP antibody and was assayed for the ability to cleave the H1t pre-mRNA when complemented with 50 ng of the indicated mutant human SLBP. Depletion of the extract resulted in an almost complete loss of processing activity (lane 2) which was fully restored when the depleted extract was supplemented with the full-length human SLBP (lane 3). Complementation of the depleted extract with the mutant proteins is shown in lanes 4 to 8. (C) Processing activity of the nuclear extract (NE; differing from that in Fig. 1B) before and after immunodepletion is shown in lanes 1 and 2, respectively. Fifty nanograms of human SLBP (lane 3) or mutant proteins was added to the depleted extract, as indicated (lanes 4 to 6). (D) The recombinant proteins were tested for their ability to bind to the stem-loop RNA oligonucleotide. Fifty nanograms of the recombinant protein indicated at the top of each lane was incubated with the radiolabeled stem-loop RNA, and the complexes were resolved by native gel electrophoresis.