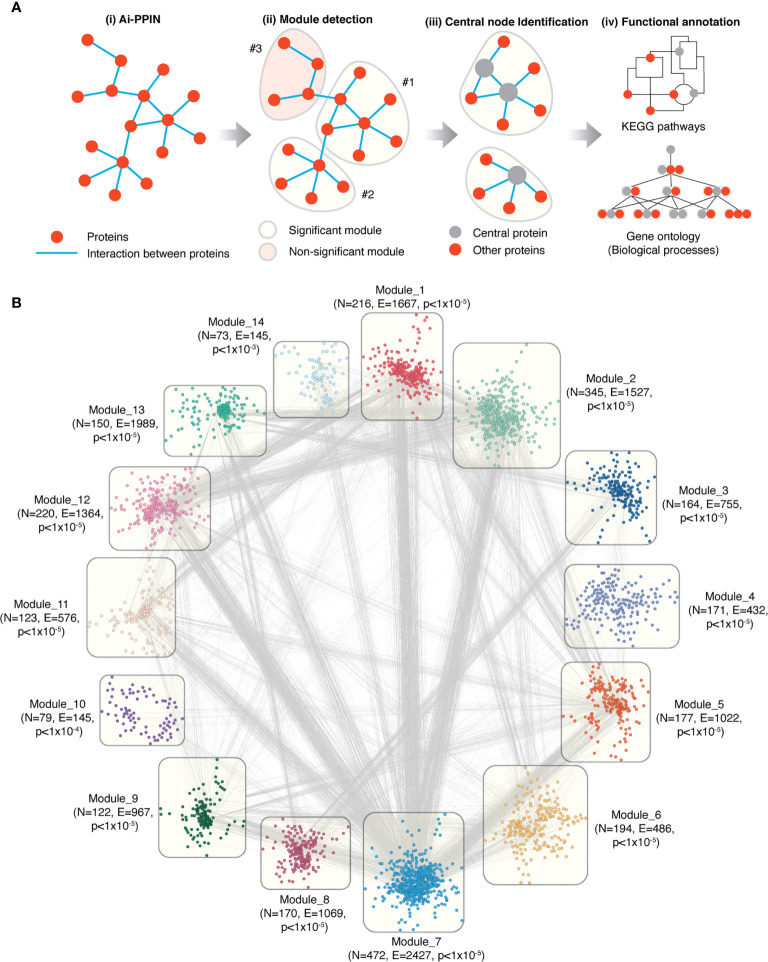
Figure 2.
Overview of the functional modules identified from Ai-PPIN. (A) Schematic representation of the Ai-PPIN module analysis. The Louvain community detection algorithm (18) was applied to detect communities/modules within the Ai-PPIN network. Statistically significant (qs-test) (21) modules (yellow bubble) were identified by comparison with modules from 10000 random networks. Non-significant modules (red bubble) were excluded from further analysis. Functional enrichment analyses using KEGG pathways and GO : BP (gene ontology biological process terms) were performed to identify the biological functions enriched within each module. (B) The Ai-PPIN contains fourteen significant modules. In each module, the nodes represent proteins. The lines connecting the nodes represent interactions between proteins. N and E denotes the number of nodes and edges present in each module respectively. The p-value denotes the statistical significance of the modules (qs-test) (21). Cytoscape (version 3.8.2) was used for visualization of the network.