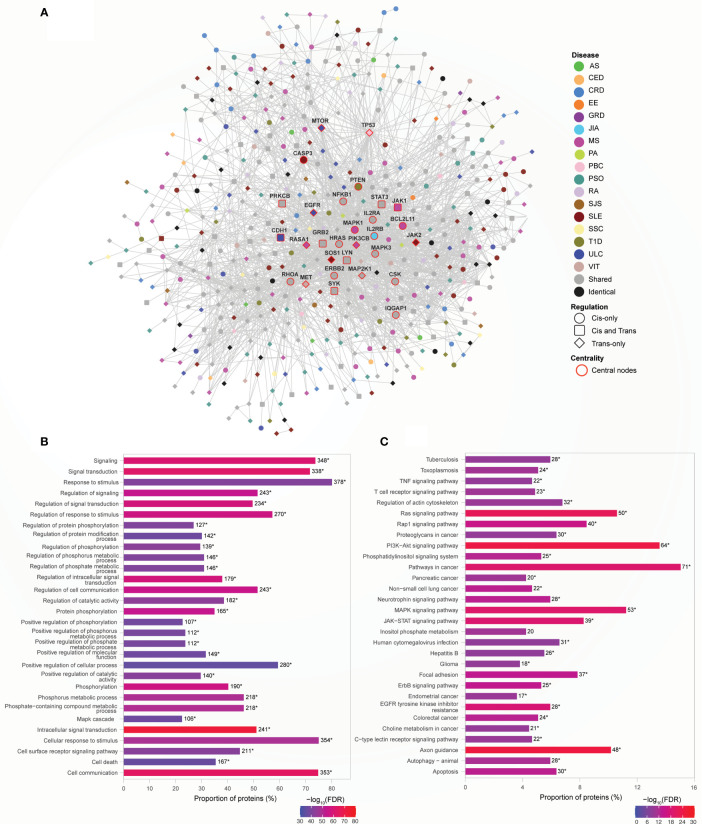
Figure 6.
The largest network module is enriched for cellular signaling and cancer pathways. (A) Network representation of the module. The color of the nodes denotes the disease with which the protein is associated. Node shape indicates if the SNP acts locally (cis - circle), distally (trans - diamond), or both (cis and trans – rounded square) on the genes encoding proteins. Central nodes are highlighted in red borders and labelled. Cytoscape (version 3.8.2) was used for visualization of the module. (B) Module 7 is enriched for signaling and metabolic processes. The top 30 enrichment results are shown (FDR ≤ 4.77E-36). (C) KEGG pathway enrichment analysis identified enrichment in signaling and cancer related pathways (FDR ≤ 3.70E-09). The top 30 pathway enrichment results are shown. In (B, C), the numbers, to the right of each bar, denote the number of proteins enriched for that term or pathway. The asterisk designates terms or pathways that were enriched for shared proteins that are central to the network (Supplementary Data 12 Tables 4, 5 respectively).