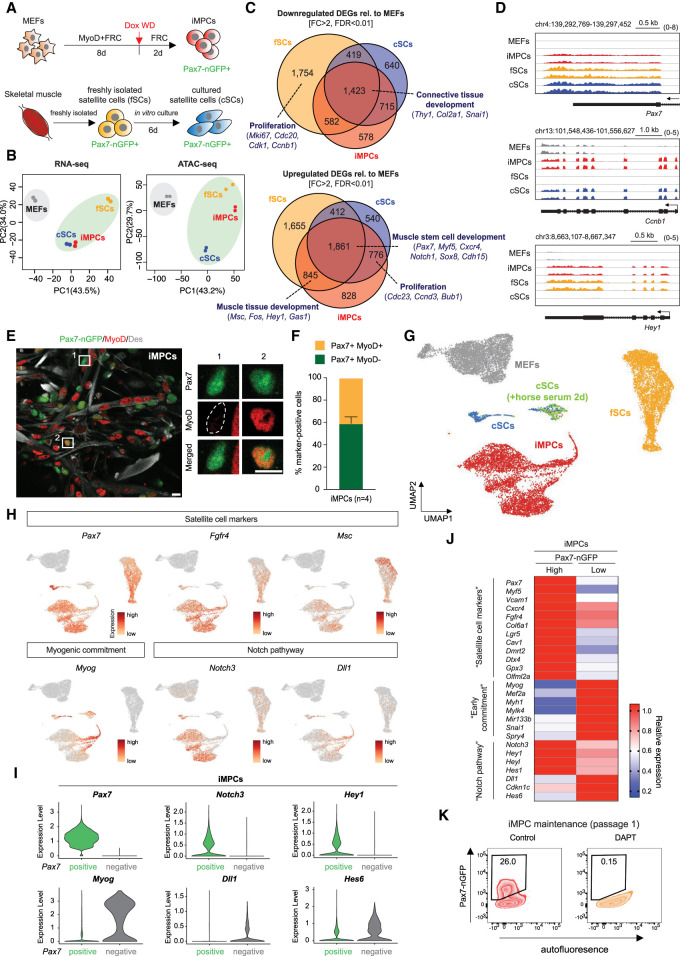
Figure 2.
Pax7+ iMPCs share key characteristics with satellite cells. (A) Experimental outline to isolate Pax7-nGFP+ fresh SCs (fSCs), cultured SCs (cSCs), and iMPCs. (Dox WD) Dox withdrawal. (B) PCA of RNA-seq and ATAC-seq data for the indicated samples (n = 2 per sample). (C) Venn diagram showing the overlap of up-regulated and down-regulated differentially expressed genes (DEGs) when comparing MEFs with either iMPCs, fSCs, or cSCs (n = 2 per sample). (D) RNA-seq gene tracks of representative myogenic genes highlighted in C. (E) Immunofluorescence images showing expression of MyoD, Des, and Pax7-nGFP in established iMPCs maintained in FRC without exogenous MyoD (Dox). Scale bar, 10 µm. (F) Quantification of Pax7+/MyoD− and Pax7+/MyoD+ iMPC subsets shown in E. Error bars indicate mean ± SD (n = 4). (G) UMAP embedding of single-cell RNA-seq data for indicated samples. (H) Expression of representative genes using the same embedding as in G. (I) Expression levels of indicated genes in Pax7-nGFP+ iMPCs with (“positive”) or without (“negative”) endogenous Pax7 mRNA signal. (J) Heat map showing differentially expressed marker genes (RNA-seq) between Pax7-nGFPhigh (n = 2) and Pax7-nGFPlow (n = 2) iMPCs. (K) Effect of DAPT (Notch inhibitor) on iMPC maintenance.