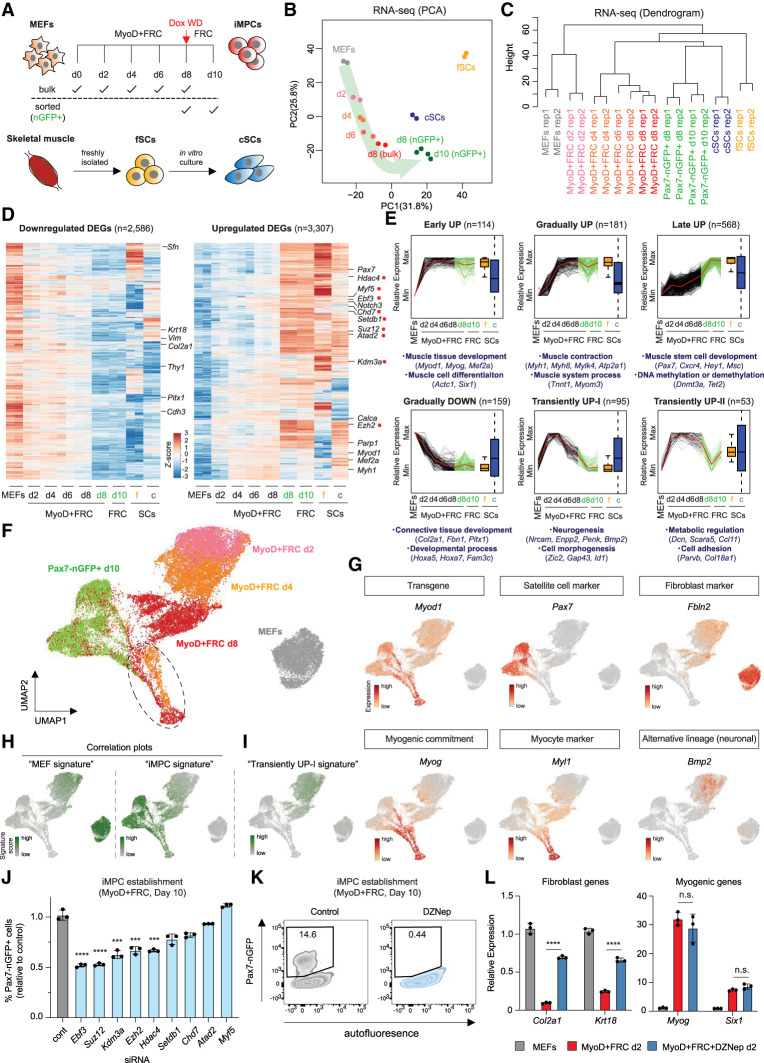
Figure 3.
PRC2-dependent, gradual silencing of the fibroblast program precedes late induction of the stem cell program. (A) Experimental outline to study transcriptional dynamics of dedifferentiation. Pax7-nGFP+ fSC, cSC, and iMPC (d10) samples are identical to those in Figure 2. (B) PCA of RNA-seq data for the indicated samples (n = 2 per sample). (C) Hierarchical clustering dendrogram of RNA-seq data for the indicated samples. (D) Heat maps of gene expression (z-scores of log2 transformed RPKM relative to all samples) for genes differentially expressed (fold change [FC] > 2, FDR < 0.01) in iMPCs (GFP+) and at least fSCs or cSCs compared with MEFs (n = 2 for each). (E) k-means clustering of temporal bulk gene expression profiles with representative GO terms and genes shown below. Gene expression was normalized and scaled from minimum to maximum among MEFs, reprogramming intermediates (d2–d8) and iMPCs. (F) UMAP embedding of single-cell RNA-seq data of MEFs, Pax7-nGFP+ iMPCs, and intermediates (Dox + FRC d2, d4, and d8). Dotted oval indicates Myog+ population common to days 4 and 8 and iMPCs. (G) Expression of representative genes using the same UMAP embedding as in F. (H) Expression of MEF signature genes (MEF pseudobulk) and iMPC signature genes (iMPC pseudobulk) using the same UMAP embedding as shown in F. (I) Expression of “transient UP (I)” gene signature using the same UMAP embedding. transient UP (I) genes (n = 95) were extracted from bulk RNA-seq data shown in E. (J) Quantification of Pax7-nGFP+ cells using MEFs exposed to MyoD + FRC and indicated siRNAs for 10 d. Targeted genes are highlighted with red dots in D. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (***) P < 0.001, (****) P < 0.0001. (K) Effect of DZNep (Ezh2 inhibitor) on iMPC maintenance. (L) Quantitative RT-PCR analysis for indicated fibroblast and myogenic markers after 2 d of exposure to MyoD + FRC with or without DZNep. Values normalized to MEFs. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (****) P < 0.0001, (n.s.) not significant.