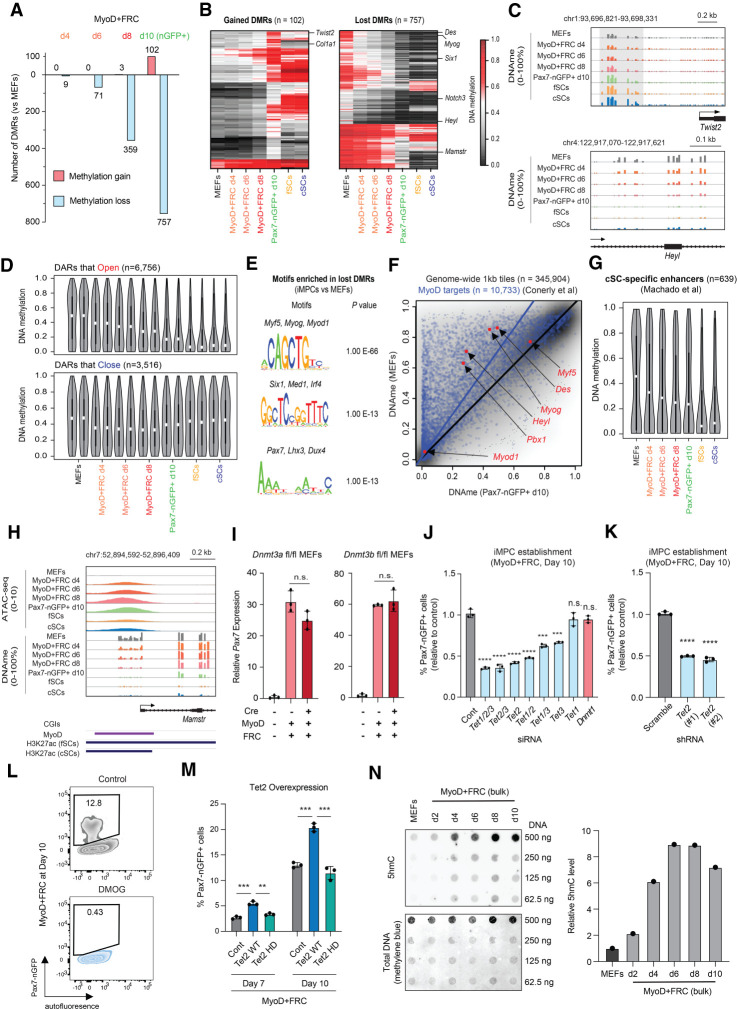
Figure 5.
Active DNA demethylation is required, while de novo methylation is dispensable for reprogramming to a Pax7+ state. (A) Number of differentially methylated regions (DMRs) that are gained or lost when comparing MEFs with the indicated time points using RRBS analysis (FDR < 0.01, CpGs > 8, and difference > 0.2). (B) Heat maps showing DMRs that are gained or lost between MEFs and iMPCs, with inclusion of fSC and cSC samples. (C) Genome browser tracks of representative DMRs shown in B. (D) Violin plots showing DNA methylation (DNAme) levels for CpGs located within DARs (differentially accessible regions) that open (n = 6756) or close (n = 3516) between MEFs and iMPCs. (E) Transcription factor motif enrichment for regions that gain accessibility and lose methylation (Pax7-nGFP+ d10 iMPCs vs. MEFs). (F) Scatter plot showing correlation between global DNAme (1-kb tiles, n = 345,904) and known MyoD targets (n = 10,733) when comparing MEFs (n = 2) with Pax7-nGFP+ iMPCs d10 (n = 2). (G) Violin plots for DNAme at CpGs located within cSC-specific enhancers (n = 639). (H) Genome browser track showing chromatin accessibility and DNAme changes at the Mamstr locus during dedifferentiation and in established iMPCs, fSCs, and cSCs. (I) Quantitative RT-PCR analysis for Pax7 in conditional Dnmt3a knockout (Dnmt3a fl/fl) or Dnmt3b knockout (Dnmt3b fl/fl) MEFs exposed to Cre and MyoD + FRC for 10 d. Values normalized to untreated control (no Cre, MyoD, or FRC). Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (n.s.) not significant. (J) Detection of Pax7-nGFP+ cells using MyoD-inducible MEFs treated with the indicated siRNAs. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (***) P < 0.001, (****) P < 0.0001, (n.s.) not significant. (K) Detection of Pax7-nGFP+ cells using MyoD-inducible MEFs transduced with two different shRNA vectors targeting Tet2 or a scrambled control. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (****) P < 0.0001. (L) Effect of DMOG (Tet inhibitor) on iMPC maintenance. (M) Detection of Pax7-nGFP+ cells using MyoD-inducible MEFs overexpressing a Tet2 WT or Tet2 HD (catalytic mutant) lentiviral construct. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (**) P < 0.01, (***) P < 0.001. (N, top) Dot blot analysis for global 5hmC levels in MEFs and dedifferentiation intermediates. (N, bottom) Values are normalized to methylene blue and MEFs (n = 1).