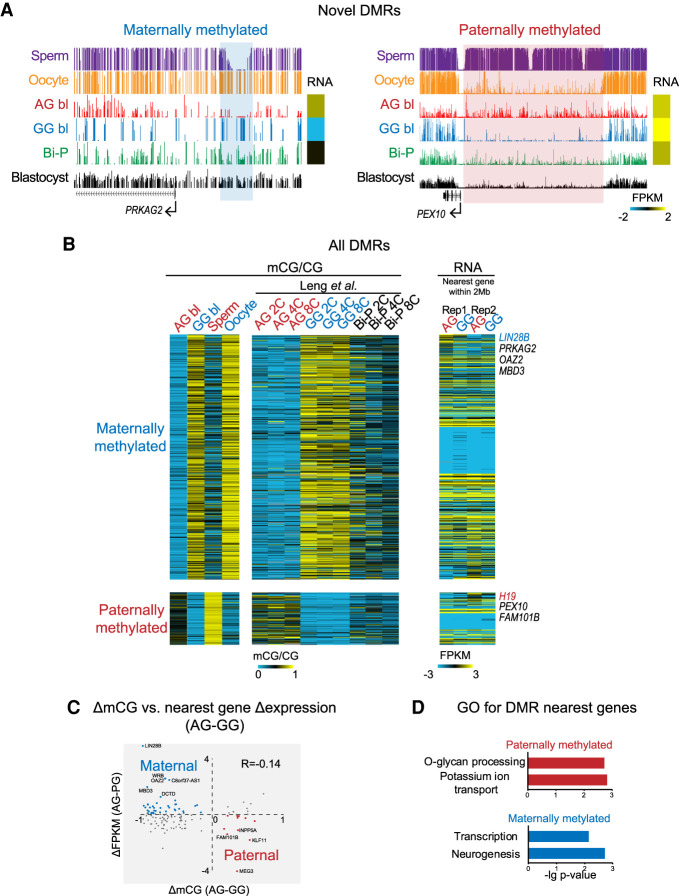
Figure 5.
Putative germline regions with differential DNA methylation. (A) UCSC snapshots showing the novel DMRs. (B) Heatmaps showing the DNA methylation levels for newly identified DMRs and the related gene expression. DNA methylation in two-, four-, and eight-cell embryos published previously was included as a control. The DMRs were firstly identified based on a pairwise comparison between AG and GG blastocysts as previously described (Zhang et al. 2018). Only those DMRs with changes in CG methylation levels between sperm and oocyte greater than 0.1 (e.g., for maternal DMRs, the oocyte showed higher methylation levels than the sperm) were taken as allelic putative DMRs (Methods). One hundred five sperm-specific DMRs and 506 oocyte-specific DMRs were detected. (C) Correlation between mCG difference and nearest gene expression difference between AG and GG blastocysts. (D) GO analysis for the nearest genes of maternal and paternal DMRs.