Abstract
Antibodies offer a powerful means to interrogate specific proteins in a complex milieu. However, antibody availability and reliability can be problematic, whereas epitope tagging can be impractical in many cases. To address these limitations, the Protein Capture Reagents Program (PCRP) generated over a thousand renewable monoclonal antibodies (mAbs) against human presumptive chromatin proteins. However, these reagents have not been widely field-tested. We therefore performed a screen to test their ability to enrich genomic regions via chromatin immunoprecipitation (ChIP) and a variety of orthogonal assays. Eight hundred eighty-seven unique antibodies against 681 unique human transcription factors (TFs) were assayed by ultra-high-resolution ChIP-exo/seq, generating approximately 1200 ChIP-exo data sets, primarily in a single pass in one cell type (K562). Subsets of PCRP mAbs were further tested in ChIP-seq, CUT&RUN, STORM super-resolution microscopy, immunoblots, and protein binding microarray (PBM) experiments. About 5% of the tested antibodies displayed high-confidence target (i.e., cognate antigen) enrichment across at least one assay and are strong candidates for additional validation. An additional 34% produced ChIP-exo data that were distinct from background and thus warrant further testing. The remaining 61% were not substantially different from background, and likely require consideration of a much broader survey of cell types and/or assay optimizations. We show and discuss the metrics and challenges to antibody validation in chromatin-based assays.
Antibodies are a critical component of a wide variety of biochemical assays. They serve as protein-specific affinity-capture and detection reagents, useful in vivo and in vitro. Example assays include chromatin immunoprecipitation (ChIP) of protein–DNA interactions, immunofluorescence, immunoblotting, ELISA, purification of cells and proteins, protein binding microarray (PBM) experiments, and targeted in vivo delivery of effector molecules (Chames et al. 2009; Park 2009; Siggers et al. 2011a; Mahmood and Yang 2012; Engelen et al. 2015; Lin et al. 2015). One advantage of target-specific antibodies is their ability to recognize proteins without the need for an engineered affinity tag. The human proteome contains tens of thousands of distinct proteins, each requiring a different antibody for specific detection. The usage of a variety of antibodies to diverse targets has been a critical component of NIH-funded consortium projects such as The ENCODE Project Consortium and Roadmap Epigenomics Mapping (The ENCODE Project Consortium 2012; Roadmap Epigenomics Consortium et al. 2015). However, broad profiling of the genomic targets of human sequence-specific transcription factors (ssTFs) has been limited by the availability of “ChIP-grade” antibodies.
Overall, there has been an acute lack of antibodies that effectively distinguish the many thousands of different chromatin proteins. Consistency in reagent production and performance has been particularly problematic (Egelhofer et al. 2011; Baker 2015; Shah et al. 2018). Polyclonal antibodies, being a mixed product of many antibody genes, have the advantage of potentially recognizing multiple epitopes on a protein, thereby producing robust target detection (Hanly et al. 1995). Large-scale efforts to generate polyclonal antibodies against human proteins, such as the Human Protein Atlas, have been successful at generating immunohistochemically competent antibodies (Uhlen et al. 2016). However, their production is finite, can be variable across immunized animals, and may also vary within individuals by different bleed dates and affinity purifications. These factors and more hamper reproducibility (Reardon 2016). Although some groups have made attempts to generate renewable immunoreagents that are viable in a wide range of biochemical assays, most efforts have been limited to a relatively small number of proteins (Colwill et al. 2011).
The NIH Protein Capture Reagent Program (PCRP) was initiated through the NIH Common Fund with the stated goal of testing the feasibility of producing low-cost, renewable, and reliable protein affinity reagents in a manner that can be scaled ultimately to the entire human proteome (PA-16-287) (Blackshaw et al. 2016; https://proteincapture.org/). With an initial focus on putative ssTFs, this endeavor reported the production of 1406 mouse monoclonal antibodies (mAbs) against 737 chromatin protein targets (Venkataraman et al. 2018). This included two parallel production approaches: mouse hybridomas that release mAb into growth medium supernatant, and recombinant antibodies produced in Escherichia coli (Hornsby et al. 2015). The advantages of these two approaches over polyclonal antibodies are, in principle, a renewable and consistent supply of homogeneous preparations produced from a single set of genes that recognize a single epitope (Köhler and Milstein 1975; Winter et al. 1994; Liu 2014). To accommodate the potential shortcoming of a hybridoma recognizing a single nonviable epitope, the NIH PCRP made an effort to generate at least two independent clones for each target, although this does not guarantee two different epitopes, as in cases in which there are immunodominant regions.
Antibody validation is required to generate confidence in their utility (Baker 2015; Marx 2019). Validation exists at many levels, ranging from whether an antibody specifically recognizes its intended target to the exclusion of all others to whether it consistently performs successfully in a particular assay (Landt et al. 2012; Wardle and Tan 2015; Uhlen et al. 2016; Edfors et al. 2018; Sikorski et al. 2018). Each publicly available PCRP-generated antibody was previously validated for its target recognition by in vitro human protein (HuProt) microarray screening (Venkataraman et al. 2018). These arrays contain approximately equivalent amounts of antigen, which differs from the wide expression range in natural sources. They also may differ in epitope accessibility compared with complexed or cross-linked targets in ChIP assays. Thus, additional assay-specific validation is necessary. The further capability of PCRP antibodies has been described for a limited set in a number of approaches, including immunoblotting, immunoprecipitation, immunohistochemistry, and ChIP-seq, with assay-dependent success reported (Venkataraman et al. 2018).
Previous work has reported that 45 of 305 mAbs against 36 of 176 targets passed ENCODE ChIP-seq standards (Venkataraman et al. 2018), although detailed supporting evidence is not publicly available. “Browser shots” of selected loci from that study are available for approximately 40 data sets against 31 targets. However, these should be considered preliminary because locus-specific examples lack statistical power and unbiased selection. Additionally, chromatin fragmentation and extraction may generate localized variation in yield that varies from prep to prep (active promoters, enhancers, etc.). Numerous replicates (target and control) are often needed to ensure against false positives at selected individual loci owing to sampling variation and multiple hypothesis testing.
As broader community use of the PCRP-generated antibodies will likely benefit from a wider survey, we conducted additional tests of these reagents. To our knowledge, there has been no systematic large-scale field assessment of antibodies in ChIP. We report on the progress and challenges in comprehensively assaying approximately 1400 PCRP mAbs. As this represents a first-pass assessment, most experiments include only a single replicate, using enrichment of specific genomic features as preliminary evidence of success. We performed replicates on some samples that displayed enrichment (e.g., expected motif enrichment), as well as a subset of samples that displayed no initial enrichment to examine our true-negative rate.
Because evaluating each of approximately 1400 mAbs in a wide variety of assays was not practical, we opted for broad coverage by ChIP-exo, which we have developed into a high-throughput and ultra-high-resolution alternative to ChIP-seq (Rossi et al. 2018). ChIP-exo allows genome-wide detection of chromatin interactions at near-base-pair resolution, which also increases the confidence of peak calling. We further tested a smaller subset of mAbs in other assays (ChIP-seq, CUT&RUN, super-resolution cellular microscopy [STORM], immunoblots, and PBMs). These additional tests were not intended to be comprehensive but rather to evaluate the challenges and practicality of systematic antibody validation. Overall, we tested 943 unique mAb clones (887 in ChIP-exo and 59 in other assays), of which 642 targeted putative ssTFs. This allowed for computational comparison through the enrichment of their cognate motifs (if one exists). The antibodies and assays were chosen to cover a wide range of end-user applications, with specific ssTFs chosen in part based on the scientific interests of the investigators and a set of objective criteria. With a deep dive on a single assay (ChIP-exo), we explored end-user practical issues related to antibody sourcing, reproducibility and validation metrics, and specificity for cell types and states.
Results
Screening PCRP mAbs by ChIP-exo
We used the massively parallel ChIP-exo version of ChIP-seq to screen PCRP Abs in a 96-well plate format (48 at a time) for their ability to recognize their putative protein targets in a chromatinized, cellular context (Rhee and Pugh 2012; Rossi et al. 2018). Briefly, proteins were formaldehyde cross-linked to DNA and each other within cells. Chromatin was then isolated, fragmented, and immunoprecipitated. While on the beads, the fragmented DNA was trimmed with a strand-specific 5′–3′ exonuclease up to the point of cross-linking (i.e., protection), which was then mapped by DNA sequencing. For many proteins, this provides single-base-pair resolution in genome-wide detection (Rhee and Pugh 2011). Because ChIP-exo is a higher-resolution derivative of ChIP-seq, ChIP-exo is expected in principle to detect any real binding events that ChIP-seq detects and likely more owing to its higher dynamic range.
Technical reproducibility of ChIP-exo with PCRP mAbs was evaluated with 43 independent replicates performed on the sequence-specific TF USF1. A USF1 ChIP replicate served as a positive control in 43 cohorts of 46 mAbs assayed on different days. No USF1 replicates were excluded in this analysis. We prioritized mAb evaluation in K562 (human bone marrow lymphoblast) but also tested a subset in MCF-7 (human mammary gland epithelial), HepG2 (human liver, epithelial-like), and donated human tissues (liver, kidney, placenta, and breast). From this and IgG (or no antibody)-negative controls, we defined with USF1 a set of approximately 164,000 E-box motif instances associated with a significant (Q < 0.01) ChIP-exo peak-pair in at least one replicate experiment (Albert et al. 2008). This reflects a very relaxed criterion, with a high level of expected false positives, for the purposes of evaluating the gradient from true binding through nonspecific background at genomic E-boxes. The latter is expected when a marginal peak location occurred in only a small fraction of data sets. When examined at greater stringency regarding the number of replicates in which the same peak was found, a higher average occupancy and more robust patterning were observed. Of the USF1 data sets, 43 of the 45 (>95%) produced a USF1-specific ChIP-exo pattern around E-boxes (Supplemental Fig. 1A, vertical blue and red stripes in the heat maps and single-base-pair peaks in the composite plots). A Pearson's pairwise correlation was calculated for the occupancy of binding at putative USF1-bound E-boxes across all USF1 and negative control IgG (or no antibody) ChIP data sets (Supplemental Fig. 1B). A strong correlation among USF1 data sets was observed, but not among the negative controls, which reflects a high level of reproducibility and indicated that ChIP-exo was suitable for screening and evaluating PCRP mAbs in ChIP.
We initiated our ChIP mAb survey by first considering the practicality of producing a large number of antibody preparations from E. coli or mouse hybridomas. Purification of these immunoreagents from E. coli included transformation of expressing plasmids, cell growth, recombinant protein induction, and purification. After multiple attempts, we determined that high-throughput parallelized recombinant immunoreagent production was not practical within the scope of this project, owing to a need to optimize the protocol in our hands. We therefore opted to pursue commercially available hybridoma-based mAbs.
We first examined vendor source. We tested NRF1, USF1, and YY1 PCRP mAbs from the Developmental Studies Hybridoma Bank (DSHB) and CDI Laboratories. The former was supplied as hybridoma culture supernatants (10–80 ug/mL antibody); the latter, as concentrates. Each preparation (as supplied) was preloaded onto protein A/G (pAG) magnetic beads, and an equal volume of each library was pooled for sequencing (to compare relative ChIP-yield). In general, we found that although mAbs from both sources (assayed at the same reported mAb amounts; 3 µg) specifically detect NRF1 and USF1, DSHB-derived hybridoma culture supernatants detected more binding events at cognate motifs compared with CDI concentrates in these ChIP-exo experiments (Supplemental Fig. 2). Based on these collective results, we sourced hybridomas from DSHB for the remainder of this study. Nevertheless, mAbs from CDI likely can be improved through further optimizations.
We assayed all 887 available hybridoma supernatants containing mAbs to 681 nonredundant targets. Testing was primarily performed in K562 (1009 data sets), although a subset of hybridoma supernatants was tested in MCF-7 (134 data sets) and HepG2 (96 data sets) cells. In the initial phase of the project, cell lines were selected based on reported target mRNA expression levels. If there was no substantial difference (less than twofold FPKM difference) in ssTF expression level or if the ssTF was not measurably expressed in any of these three lines, testing defaulted to K562 (Supplemental Fig. 3A). Later stages of validation prioritized K562 as the sole source of validation owing to practical considerations, including the ability to grow this cell line at scale (liquid culture). Two hundred forty-five unique hybridoma clones were assayed in replicate at least twice in the same or different cell types, resulting in 1261 data sets (Supplemental Fig. 3B). Of these 245 hybridoma clones, 36 (14.7%) showed enrichment of the same class of genomic features. One hundred two (41.6%) of the mAb clones produced no enrichment of genomic features in both replicates, and 107 (43.7%) produced enrichment in one sample but not in the other (Supplemental Table 2). The latter may be owing to being at the limits of detection. We next set out to characterize certain mAb in more depth.
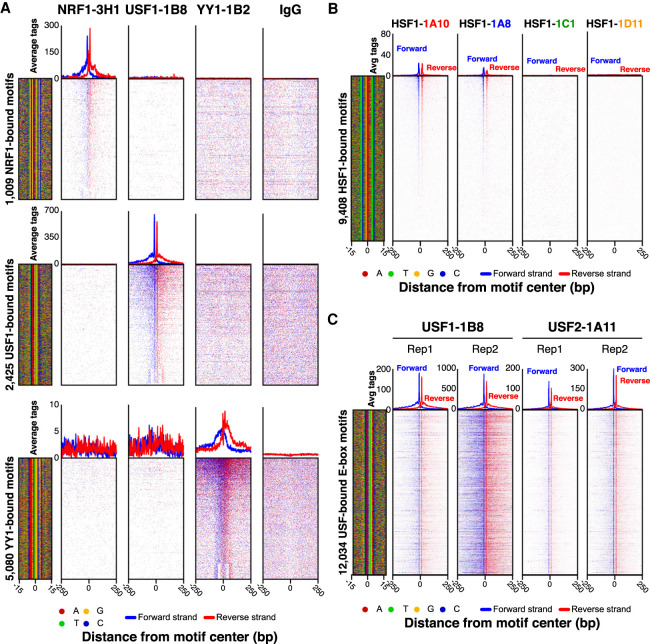
As exemplified by NRF1, USF1, YY1, and an IgG-negative control (Fig. 1A), the finding that ChIP-exo peaks were enriched at a very precise distance from cognate motifs provided strong support for specificity in target detection. We also compared different hybridoma clones against the same target, as exemplified by heat shock transcription factor 1 (HSF1). Hybridoma clones potentially target different epitopes, although immunodominance may yield independent clones to the same epitope. Both HSF1 mAbs gave nearly identical ChIP-exo read patterns around the same set of features (heat shock elements) (Fig. 1B, clones 1A10 and 1A8), thereby showing reproducibility of ChIP-exo profiles across independent PCRP mAbs. This is particularly important where validation criteria by motif enrichment are not applicable. However, two other HSF1 mAb clones failed (Fig. 1B, clones 1C1 and 1D11), indicating that independent PCRP clones can have different capabilities in ChIP. Therefore, if one mAb clone fails, it may be productive to check others.
Figure 1.
Validation of PCRP mAbs in ChIP-exo. (A) Comparison of ChIP-exo data at cognate versus noncognate motifs. ChIP-exo heatmap, composite, and DNA-sequence four-color plots were generated for NRF1, USF1, YY1, and IgG ChIP-exo data sets against the complete matrix of bound motifs from Supplemental Figure 2. The 5′ end of aligned sequence reads for each set of experiments was plotted relative to the distance from the cognate motif for each indicated target. Reads are strand-separated (blue, motif strand; red, opposite strand) and total-tag-normalized across samples. Rows are linked across samples and sorted based on their combined average rank-order in a 100-bp bin around each motif midpoint. High levels of background result in a more uniform distribution of reads across the window (as seen with the IgG control). (B) Enrichment comparisons of four unique HSF1 hybridoma clones (HSF1-1A10, HSF1-1A8, HSF1-1C1, HSF1-1D11). ChIP-exo heatmap, composite, and DNA-sequence four-color plots are shown for the indicated number and type of bound motifs for the indicated antibody hybridoma clones (A,B) or interaction partners (C) tested in K562 cells. The 5′ end of aligned sequence reads for each set of experiments was plotted against the distance from the cognate motif, present in the union of all called peaks between the data sets for each indicated target. Reads are strand-separated (blue, motif strand; red, opposite strand) and total-tag-normalized across samples. Rows are linked across samples and sorted based on their combined average rank-order in a 100-bp bin around each motif midpoint.
Targets that interact with each other or with the same sites may also provide a useful validation criterion for determining enrichment specificity. For example, in the case of USF1 and USF2 interaction partners and homologs (Rada-Iglesias et al. 2008), the USF1-1B8 and USF2-1A11 mAbs detected binding at the same sites (Fig. 1C). However, in this particular case, we cannot exclude cross-reactivity of the mAb with the two homologous USF1/2 proteins (always a potential concern with target-specific antibodies). Additional validation criteria may include comparisons to public-domain ChIP-seq data sets that use different antibodies (e.g., ENCODE, as in Supplemental Fig. 4).
Assessment by ChIP-seq
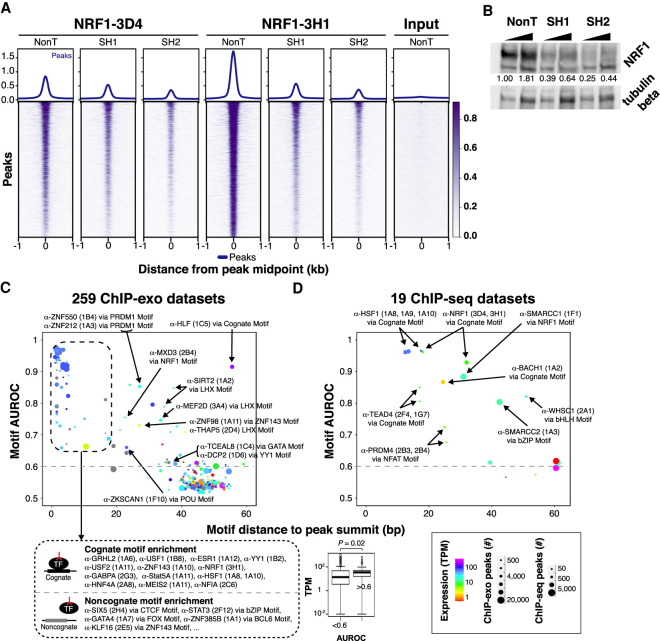
Because ChIP-seq is a widely used assay (and related to ChIP-exo), we performed ChIP-seq (in HCT116 cells) with 137 PCRP hybridomas corresponding to 70 targets associated with chromatin binding, modification, enhancer function, and/or transcriptional elongation. We found 19 (14%) produced significantly enriched peaks (see Methods) (Supplemental Table 3). However, these single-replicate data sets were not checked for enrichment of specific classes of genomic features. Stringent validation for antibody specificity involves knocking down a target and then observing a reduction of assayed signal relative to a mock knockdown (Wardle and Tan 2015; Uhlen et al. 2016; Edfors et al. 2018). We examined the feasibility of this starting with one target, NRF1. NRF1 expression was knocked down in HCT116 cells by RNAi. NRF1 peaks were concomitantly diminished with two different specific oligos but not by an untargeted oligo (Fig. 2A), thereby showing specificity of the PCRP mAbs 3D4 and 3H1 for this target. We further confirmed specific knockdown of NRF1 by immunoblot (Fig. 2B). Because of the relatively high cost and current limitations in knockdown technologies, we found it was not practical within the scope of this project to conduct systematic knockdowns across the PCRP mAb collection. Furthermore, knockdown validation may not provide the level of validation stringency in ChIP that it does for immunoblots. Knockdown of proteins can cause widespread indirect effects on the binding of other protein complexes, which could in turn skew the ChIP-signal in aberrant ways (Trescher and Leser 2019).
Figure 2.
Examining specificity of PCRP mAbs. (A) Heatmaps and composite plots displaying the global loss of NRF1-3D4 and NRF1-3H1 ChIP-seq signal after NRF1 RNAi. HCT116 cells were treated with nontargeting (sh Control) or two different NRF1-directed shRNAs (shRNA 1 and shRNA 2). Rows are linked across samples and sorted in descending order by mean score per region. (B) Western blot analysis of NRF1 knockdown by two different shRNAs (SH1 and SH2) or a nontargeting shRNA (NonT). HCT116 cells were infected with the indicated shRNAs and selected with puromycin (2 μg/mL). Total cell extracts were prepared for SDS-PAGE and immunoblotting against NRF1 and tubulin beta as the loading control. NRF1 knockdown efficiency (upper band in top panel) was quantified after normalizing with tubulin beta levels using ImageJ, and the normalized values shown. (C) Motif enrichment analysis of ChIP-exo. Cartoons depict models for binding via the cognate motif of the target ssTF or noncognate binding. Box plots of TPM expression values of target ssTFs associated to antibodies stratified by AUROC value. Results from analysis of 100 putative ssTF binding motifs within each ChIP-exo data set with more than 500 peaks (259 data sets in total). We assigned to each ChIP-exo data set the PWM with the highest AUROC (“top motif”) and quantified its centering as the mean distance of the PWM match from the peak's summit. In the scatter plot, each point represents the enrichment/centering of the top motif in one of the 259 putative TF ChIP-exo data sets. Colors indicate the expression level (RNA-seq TPM value; unavailable values are shown in gray) (The ENCODE Project Consortium 2012) of the gene specific for the antibody used in the ChIP assay. Point sizes indicate the number of ChIP-exo peaks in the data set. Top motifs with AUROC > 0.6 (dashed line) and TPM values from duplicate RNA-seq experiments are indicated. (D) Results from enrichment analysis of 100 TF binding motifs within each of 19 ChIP-seq data sets. Points are formatted as in C.
Assessment through feature enrichment
Thus far, we have established the utility of three independent validation criteria inherent to ChIP-exo analysis: (1) enrichment and patterning at a cognate motif (Fig. 1A), (2) correlation with an independent mAb clone (Fig. 1B), and (3) colocalization with an interacting partner (Fig. 1C). In our large-scale evaluation of mAbs, these validation criteria often were either not applicable or not attainable. We therefore looked for additional criteria that might be useful where the preferred validation criteria were inconclusive. We used the ChExMix algorithm to identify significant modes of protein binding and de novo motif detection through a combination of DNA sequence enrichment and variation in ChIP-exo patterning (Yamada et al. 2019). Discovered motifs were identified using TOMTOM and the JASPAR database (Gupta et al. 2007; Fornes et al. 2020). The relative enrichment of peaks in annotated genomic regions (ChromHMM and Segway genome segmentations) was quantified (Supplemental Fig. 3A; Hoffman et al. 2013). We also considered a low stringency test that did not require statistical enrichment of peaks. Composite plots were generated around transcription start sites (TSSs) and CTCF binding sites, and the average tag enrichment was examined relative to a negative control (IgG) background. Test results for all validation criteria and other analyses for each tested antibody can be found at www.PCRPvalidation.org and Supplemental Table 2. The website provides a deep and rich resource for preliminary discovery for each target, particularly because many of targets remain uncharacterized. We caution that the website provides automated analysis for all data sets, including those that did not pass our significance thresholds and/or were not replicated independently. The analyses should serve only as a reference point for additional characterization and optimization and should not be used to draw biological conclusions.
Evaluation through motif analysis
To further evaluate the ChIP-exo and ChIP-seq data for evidence of ssTF genomic occupancy (direct or indirect), we analyzed 259 ChIP-exo and 19 ChIP-seq peak files for ssTF motif enrichment by using an area under the receiving operator characteristic curve (AUROC) metric in which ChIP “bound” regions were compared to a background set of unbound sequences. Briefly, the AUROC assesses the enrichment of matches to a given TF motif among the ChIP “bound” regions compared with a background set of unbound regions; the resulting AUROC value ranges from zero to one, with 0.5 corresponding to that expected at random. In addition to AUROC motif enrichment, we also quantified the distance of the motif to the peak summit, which is expected to be shorter for motifs recruiting the profiled ssTF to the DNA (either directly or through a tethering ssTF partner) (Fig. 2C,D; Gordan et al. 2009; Bailey and Machanick 2012; Wang et al. 2012; Mariani et al. 2017). We used a collection of 100 nonredundant position weight matrices (PWMs) representative of the known repertoire of human ssTF binding specificity (Bailey and Machanick 2012). These approaches identified 20 PCRP antibodies, corresponding to 16 putative ssTFs, for which their cognate DNA motif was both enriched and centered within the ChIP peaks (“Direct Binding” in Supplemental Table 4). Thirty PCRP mAbs showed enrichment for a binding motif other than the cognate motif of the ChIP-profiled ssTF. Possible reasons for the remaining data sets not showing significant motif enrichment include (but are not limited to): (1) the target TF was not expressed at sufficiently high levels or at sufficiently high nuclear concentrations in the assayed cells, (2) the epitope recognized by the antibody was not accessible in the chromatin context in the assayed cells, (3) the target TF was not occupying specific genomic target sites (either directly or indirectly) in the assayed cells, or (4) there was off-target recognition by the antibody of other proteins in the assayed cells, resulting in lack of sufficient enrichment of the intended target TF.
ChIP assessment in multiple cell states and types
Any number of targets may be sequestered in a state that prevents their interaction with chromatin (and thus detection by ChIP) unless activated to do so through a change in cell state. We examined this with HSF1, which is rapidly induced to bind in the nucleus upon heat shock to activate heat shock–response genes (Baler et al. 1993). HSF1 was bound to cognate motifs at relatively low levels under nonstressed conditions but increased in binding upon treatment of cells with hydrogen peroxide (0.3 mM) for 3 min and 30 min (assayed by ChIP-exo) (Supplemental Fig. 5A) or upon heat shock (shift from 37°C to 42°C for 1 h; assayed by ChIP-seq) (Supplemental Fig. 5B). This illustrates a potential problem with using mRNA expression levels as a basis for expecting a factor to be actively bound to chromatin. Many TFs are sequestered and only bind chromatin when released by signaling events.
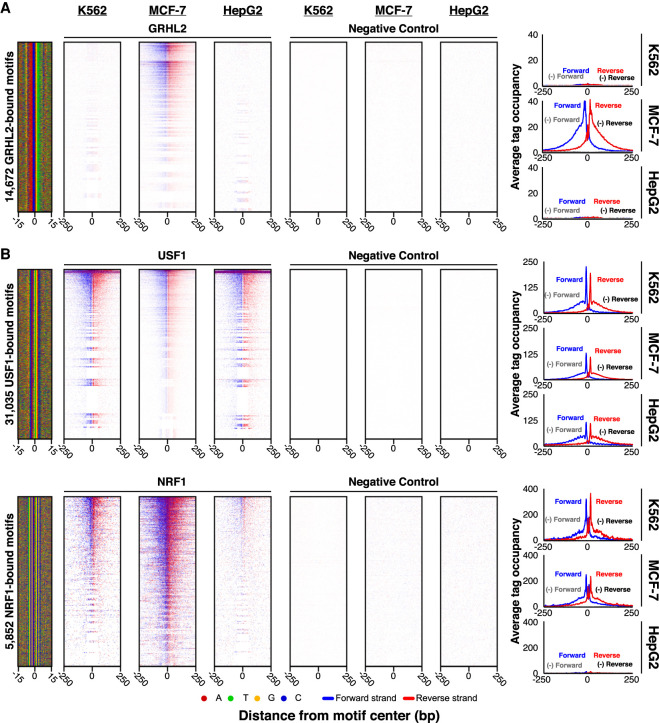
Although ChIP-exo antibody assessments were primarily performed in K562 cells (see above), many targets may have chromatin interactions that are cell type specific. An example of cell type–specific expression was observed with the breast cancer factor GRHL2, where binding was detected in MCF-7 cells but not in other cell types (Fig. 3A). Other targets like USF1 and NRF1 were less cell type specific, although we do not exclude selectivity at subsets of sites (Fig. 3B). Therefore, testing antibodies in the appropriate cell type (with appropriate signaling events) may be critical for target detection.
Figure 3.
Cell type comparison of antibody performance. (A,B) ChIP-exo heatmap, composite, and DNA-sequence four-color plots are shown for the indicated number of bound motifs for the indicated targets, in the indicated cell types. The 5′ end of aligned sequence reads for each set of experiments was plotted against the distance from the cognate motif, present in the union of all called peaks among the data sets for each indicated target. Reads are strand-separated (blue, motif strand; red, opposite strand) and total-tag-normalized across samples. Rows are linked across samples and sorted based on their combined average in a 100-bp bin around each motif midpoint.
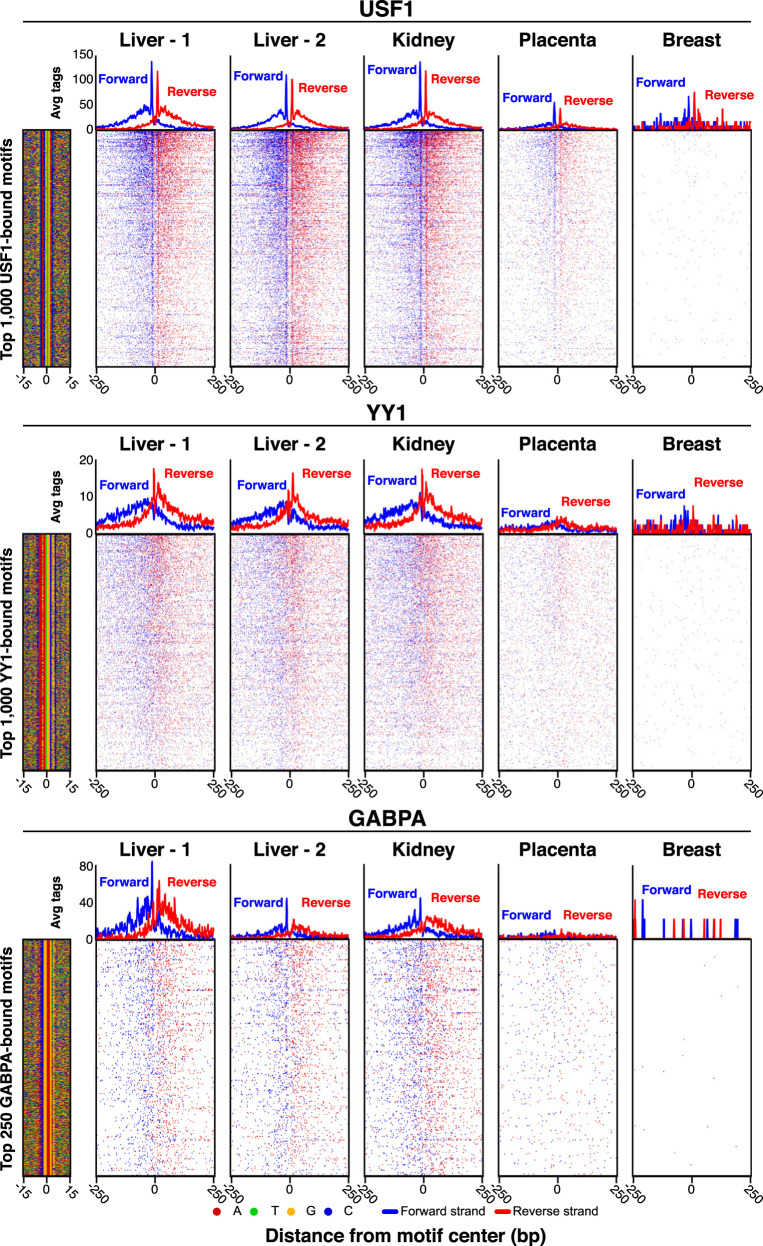
We next tested a subset of PCRP mAbs on donated deidentified human organs. ChIP-exo was performed using mAbs against USF1, YY1, and GABPA in chromatin from human liver (two different specimens), kidney, placenta, and breast tissue (Fig. 4). Largely consistent with results obtained in cell lines, de novo peak enrichment at cognate motifs and their aligned read patterning was observed with all three mAbs for the liver and kidney. Peak enrichment was diminished in the placenta and was not detectable in the breast. It remains to be determined whether the lack of signal in the breast is owing to technical limitations in chromatin yields versus tissue specificity of chromatin interactions. Nonetheless, these findings show the utility of at least some PCRP mAb in epigenomic profiling of human clinical specimens.
Figure 4.
Application of ChIP-exo in human tissue using PCRP mAbs. ChIP-exo heatmap, composite, and DNA sequence four-color plots are shown for the indicated number and type of bound motifs for the indicated targets, in the indicated organ types (the liver includes two donors). The 5′ end of aligned sequence reads for each set of experiments was plotted against the distance from the cognate motif, present in the union of all called peaks between the data sets for each indicated target. Reads are strand-separated (blue, motif strand; red, opposite strand) and total-tag-normalized across samples. Rows are linked across samples and sorted based on their combined average in a 100-bp bin around each motif midpoint.
Evaluation using CUT&RUN
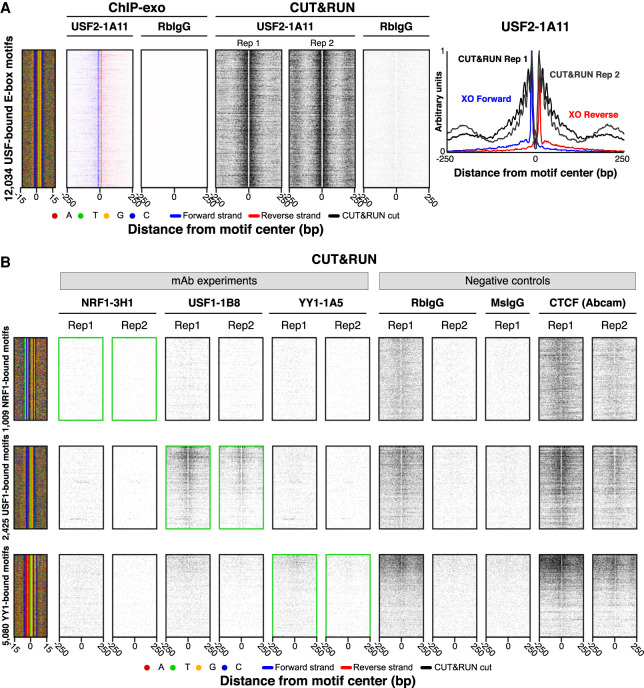
CUT&RUN has been used to measure genome-wide protein–DNA interactions (Skene et al. 2018). It uses a fusion of pAG (which binds most antibody isotypes in common use) and micrococcal nuclease (MNase). A ssTF-specific antibody is added to immobilized permeabilized cells or nuclei (under native or cross-linked conditions), where it binds to its chromatin target. pAG-MNase is next added and recruited via pAG to the target-specific antibody, and the MNase portion cleaves local DNA. The result is a selective release of chromatin from the otherwise insoluble nucleus, where genomic enrichment can be identified by sequencing. We tested 40 PCRP antibodies in K562 by native CUT&RUN in replicate (see Methods), of which 25 had been selected based on ChIP-exo enrichment. For USF2, NRF1, USF1, and YY1, we mapped CUT&RUN cleavage sites around their ChIP-exo detected cognate motifs. Multiple nonspecific IgGs served as the negative controls. As additional negative controls, we mapped DNA cleavages around this same set of motifs using the other noncognate ssTF CUT&RUN data sets, where only background cleavage is expected (as we did for ChIP-exo in Fig. 1 and ChIP-seq in Supplemental Fig. 4). Of all 40 PCRP mAbs antibodies tested, USF2-1A11 produced the most robust CUT&RUN signal (Fig. 5A), with a detection level matching ChIP-exo, and low IgG-only background. Thus, the native CUT&RUN assay as implemented here can detect site-specific protein–DNA interactions at ChIP-exo-validated sites through at least one PCRP mAb. However, for the NRF1, USF1, and YY1 mAbs, which had worked well in ChIP-exo, we observed little or no enrichment above background in native CUT&RUN (Fig. 5B). This may reflect intrinsic target incompatibility with the native approach, or antibody-specific optimization is needed. Analysis results for the remaining CUT&RUN data sets and controls are in Supplemental Table 5. Of note, a DNA-accessibility footprint was observed in some negative control experiments, including CTCF CUT&RUN data at NRF1, USF1, and YY1 motifs (Fig. 5B). This may indicate background cleavages by untargeted MNase fusions, which are most evident at TF binding sites. This may be because of the open chromatin at these locations and/or an intrinsic MNase sequence bias, which shows the importance of control comparisons. Background cleavage intensity may also vary based on the amount and type of IgG used in the negative control, making peak-calling less reliable. Thus, cognate target specificity in most of these CUT&RUN experiments was not established.
Figure 5.
PCRP mAb assayed by CUT&RUN. (A,B) CUT&RUN heatmap, composite, and DNA-sequence four-color plots are shown relative to the motifs defined and sorted in Figure 2. The 5′ end of aligned sequence reads is plotted. Reads are strand-separated (blue, motif strand; red, opposite strand) for ChIP-exo and combined (black) for CUT&RUN. Reads are aligned as above (Fig. 1A), and individual data set results are available in Supplemental Table 5.
Evaluation by STORM
As part of our PCRP mAb evaluation, we performed Stochastic Optical Reconstruction Microscopy (STORM), which can visualize cellular structures/processes at nanometer resolution (Betzig et al. 2006). The approach involves the use of fluorescently conjugated antibodies that might be expected to bind identifiable structures in specific subcellular compartments. Of the 39 PCRP mAbs surveyed (Supplemental Table 6), most displayed peri-cytoplasmic staining, rather than the expected punctate nuclear staining (Supplemental Fig. 6). Thus, without further supporting evidence, these results were inconclusive. We provide these images as comparison data sets for further studies in evaluating the utility of PCRP antibodies in STORM.
Evaluation using in vitro binding assays
We next tested 44 PCRP mAbs by in vitro protein binding assays. A classic method to evaluate antibody specificity is western blotting: size separation of complex protein mixtures using denaturing gel electrophoresis (SDS-PAGE), followed by membrane transfer and immunoprobing with an antibody of interest to determine the protein species it detects. Because endogenous targets can exist at a level below the sensitivity of detection, we used coupled in vitro transcription/translation (IVT) in crude HeLa cell extracts to produce 32 TFs as unpurified amino-terminal GST-fusion proteins (Supplemental Tables 7, 9). This allowed for production of higher levels of target proteins, but within a complex milieu of other proteins to allow specificity to be addressed. Of the 44 PCRP mAbs assayed by immunoblotting, 31 (70%) mAbs detected a single predominant band of the expected molecular weight (12 as biologically independent replicates of the same antibody, nine replicated with a different mAb, and 10 performed as a single replicate) (Supplemental Fig. 7). As a positive control, anti-GST antibody detected 32 of the 33 GST-fusion TFs. Thus, about two-thirds of the assayed PCRP mAbs were specific in recognizing their target proteins. This success rate (70%) may represent the upper limit of success for these reagents.
PBM is a technique to assay protein–DNA binding specificity in vitro (Mukherjee et al. 2004; Berger et al. 2006; Siggers et al. 2011b). Proteins used in PBMs are typically expressed as epitope tag fusions, supporting detection on the DNA array by fluorescent antitag antibody. A possible explanation for why some antibodies may fail to work in ChIP experiments is that their target epitope may become inaccessible when the ssTF is bound to a protein partner or DNA ligand and/or is subjected to modification by formaldehyde. Therefore, for a set of 31 ssTFs that were of interest or that performed well in ChIP, we used PBMs to test 44 PCRP mAbs for their ability to recognize their DNA-bound target TF.
Briefly, the relevant IVT-generated TFs were incubated with DNA microarrays where all possible 10-bp sequences were represented within approximately 44,000 60-bp probes on double-stranded oligonucleotide arrays (Agilent) (Berger et al. 2006). For 20 of these 44 PCRP mAbs (45%) assayed against 16 of 22 tested targets, the PBM experiments successfully identified a DNA-binding motif consistent with the known or anticipated element (Fig. 6). All mAb PBM experiments were run with a parallel anti-GST antibody (positive control) to validate the viability of the IVT-expressed target in PBMs, resulting in a 21/22 (95%) validation rate. Of the 20 PCRP antibodies that successfully yielded the expected motif in PBMs, 11 (55%) had at least some validation support by ChIP.
Figure 6.
TF binding motifs derived from PBM experiments performed using anti-GST or PCRP antibodies. Each full-length TF was assayed with its PCRP mAb(s) and compared against its corresponding motif derived from an anti-GST PBM experiment and anticipated motif from the UniPROBE or Cis-BP databases (Weirauch et al. 2014; Hume et al. 2015). The TFs from UniPROBE or Cis-BP were assayed as extended DNA-binding domains. For the display of sequence motifs, probability matrices were trimmed from left and right until two consecutive positions with information content of 0.3 or greater were encountered, and logos were generated from the resulting trimmed matrices using enoLOGOS (Workman et al. 2005).
Discussion
The ability to interrogate the diverse human proteome is heavily reliant on specific affinity capture reagents, of which antibodies are the most widely used. PCRP represented a pilot project for the entire human proteome, with initial focus on nuclear proteins. To this end, this study assayed nearly all PCRP mAb against approximately 700 putative chromatin targets or TFs using the genome-wide high-resolution ChIP-exo assay. Smaller subsets were analyzed by other assays, including ChIP-seq, CUT&RUN, STORM, immunoblotting, and PBMs. Our purpose was to present a technical “field” assessment of PCRP mAb utility in biochemical and cellular assays. Given the published rigorous criteria for antibody validation (Landt et al. 2012; Wardle and Tan 2015; Uhlen et al. 2016; Edfors et al. 2018; Sikorski et al. 2018), which may be assay specific, this work was not intended to provide a comprehensive resource of validated antibodies. Instead, it is a starting point for considering validation criteria and its limits that may be applicable to particular assays, especially when taking a systematic high-throughput approach. ChIP-exo identified up to 5% of the approximately 1000 tested PCRP antibodies as having high specificity for their targets, based on orthogonal evidence of motif enrichment and other criteria. These reagents would be the strongest candidates for more rigorous validation testing.
In contrast, many PCRP mAbs did not meet the stringent validation criteria we used. We suggest that some ambiguous outcomes would benefit from assay optimization: for example, a different cell type or media condition in which the target is expressed and/or activated for chromatin binding. Additionally, different metrics or validity thresholds may be needed. Notably, many of the PCRP mAbs were evaluated by ChIP-exo in K562. Their cognate TFs may not be appreciably expressed in K562 cells. However, as shown for HSF1, even where a target is expressed, it may not substantially interact with chromatin (and thus escape detection) unless activated to do so. Therefore, knowledge of the underlying biology of the target may be critical in how ChIP specificity is assessed.
Several algorithmic explanations can be considered for any missed target detection. Some sequence-specific DNA-binding proteins may not have been accommodated within our discovery framework. For example, a target protein may bind a nonstandard distribution DNA sequences that were not captured by current motif discovery algorithms. Alternatively, the target may interact with a wide range of genomic sites having different or degenerate DNA sequence motifs (including indirect sequence readout based on DNA shape) (Rohs et al. 2009) that are not accommodated by the discovery algorithms used in this study. Another possible scenario is that a target protein might not bind DNA directly but only indirectly through other proteins (Wang et al. 2012; Mariani et al. 2017), including those that potentially form an undiscovered chromatin class (which would not be in our discovery pipeline). The enrichment of a noncognate motif suggests that the genomic occupancy of the ChIP-profiled ssTF might be mediated through indirect binding by a different ssTF, which is bound directly to those ChIP “bound” genomic sites through the enriched motif (“Indirect Binding” in Supplemental Table 4).
We have previously identified indirect binding modes from ChIP-chip or ChIP-seq experiments that used traditionally prepared antibodies against human ssTFs or an epitope tag on yeast ssTFs (Gordan et al. 2009; Mariani et al. 2017). Here, for example, the NFAT motif was enriched and centered among ChIP-seq peaks resulting from ChIP-seq experiments using two different anti-PRDM4 PCRP mAb clones (2B3 and 2B4), suggesting that in HCT116 cells, PRDM4 could bind DNA indirectly via an NFAT factor. Another PRDM4 mAb clone (2D1) had a much smaller number of peaks that were centered around the known PRDM4 motif (Bogani et al. 2013). These peaks were also found in the ChIP-seq data of mAb 2B3 and 2B4, raising the possibility that there are two modes of binding of PRDM4 to chromatin in HCT116 cells.
We often found motifs that were long, simple, semirepetitive, and highly degenerate (see http://www.pcrpvalidation.org). These are not typical properties of sequence-specific DNA-binding proteins, and the ChIP-exo patterns at these motifs were often quite distinct from well-validated targets. Historically, some of these locations may have been set aside as problematic and thus excluded from analysis (The ENCODE Project Consortium 2012). Whether these regions are artifactual or have some unknown biology remains to be determined. Although we accepted these motifs as evidence of enrichment, we urge caution when interpreting such atypical binding events.
It was not practical in our high-throughput ChIP-exo screen to profile each PCRP mAb in a wide range of cell types. However, for ssTFs with at least 500 significant peaks, we noticed an association between the expression of the target proteins and motif detection (Fig. 2D, box plot), and so expression may be a useful preliminary guide for cell type selection. Furthermore, some targets may simply not be cross-linkable to chromatin in the assayed cell type (or any cell type), making ChIP an inappropriate assay. Unlike engineered epitope tags, each target-specific antibody may have a substantially different affinity for its cognate antigen. Therefore, we cannot rule out that at least some low-performing or nonperforming antibodies could perform better under different immunoprecipitation conditions. Still other potential reasons for antibody nonperformance may be owing to trivial explanations like lot expiration or mislabeling along the supply chain.
In total, 943 unique hybridoma clones were tested in at least one assay. We identified 50 clones (5%) that worked with high confidence in at least one of these assays. However, only a very small portion of the validation spectrum has been explored. Using relaxed criteria that may reflect significant but off-target or unknown behavior, we find that 371 (39%) of the tested PCRP mAb had at least some evidence of being different from background, in at least one assay. However, such marginal criteria require deeper characterization, such as target depletion/deletion or negative control cell lines, for a more robust validation. The remaining 61% also warrant more testing in other cell types and conditions. Compared with Venkataraman et al. (2018), we found 13/25 (52%) clones were tested and validated by ChIP by both groups (Supplemental Table 1). Differences in validation may be due in part to different cell lines, technical variability, and differences in bioinformatic criteria for validation. Our analysis identifies an initial set of prioritized candidates. A detailed summary of each assay's results along with all of the measured quality control metrics is available in Supplemental Table 1, along with an interactive searchable web-interface online at www.PCRPvalidation.org.
Methods
ChIP protocols
Antibodies
One thousand three hundred eight TF hybridomas were reported through the PCRP portal at the start of this study (September 2017). Hybridoma supernatants were purchased from Developmental Studies Hybridoma Bank (DSHB; University of Iowa) as 1 mL aliquots. mAb concentration averaged 36 ug/mL by ELISA quantification. Hybridoma supernatants contain ADCF-MAb cell culture medium (https://dshb.biology.uiowa.edu/tech-info) and residual (2%) fetal bovine serum, which has a reported IgG concentration of 1–6 ug/mL (Son et al. 2001). DSHB preparation dates were provided. Concentrated mAbs and their concentration were generously provided by CDI Laboratories.
Cell material
Cell stocks were obtained by the Pugh laboratory from ATCC. K562 were grown in suspension using IMDM media and periodically checked for mycoplasma contamination. HepG2 and MCF-7 were grown as adherent cells in DMEM media. MCF-7 cells were additionally grown in phenol red-free DMEM and treated with beta-estradiol 30 min before cell harvest. Cells were pelleted, resuspended in PBS, cross-linked with 1% formaldehyde for 10 min, and then quenched with a molar excess of glycine. Donated human organs were obtained from NDRI (Philadelphia) and then cryoground to fine powder using the SPEX cryomill cyrogrinder. Frozen tissue powder was resuspended in room-temperature PBS containing formaldehyde to a final concentration of 1% and quenched with a molar excess of glycine. All cells and tissue for ChIP-exo then proceeded through the standard lysis and sonication protocol described below after cross-link quenching. HCT116 cells (ATCC CCL-247) were grown in the Shilatifard laboratory in DMEM supplemented with 10% FBS (Thermo Fisher Scientific 35-015-CV). Seventy percent to 80% confluent HCT116 cells were heat shocked for 1 h by adding preheated conditioned media preheated to 42°C (Lim et al. 2017). Heat shock and non–heat shock HCT116 cells were washed with PBS before fixing with 1% formaldehyde (Sigma-Aldrich 252549) in PBS for 15 min and processing for ChIP-seq.
ChIP-exo testing was initially prioritized in K562, MCF-7, and HepG2, using gene expression values (FPKM in RNA-seq) generated from the ENCODE Project Consortium as the basis for the cell type used (Uhlén et al. 2005; The ENCODE Project Consortium 2012). Targets were assigned to the cell type most likely to express the protein of interest. If no cell line had a clear high expression for a specific target (>25% FPKM relative to all other considered cell lines), testing defaulted to K562. K562 was selected as the default owing to its status as a Tier 1 ENCODE cell line and the plethora of existing genomic data that could orthogonally support any findings. Samples were processed in batches of 48 in 96-well plate format. The PCRP-derived USF1 or NRF1 antibody served as a positive control for every processed cohort as well as an IgG or “no antibody” mock ChIP-negative control. Cross-linked sonicated chromatin from approximately 7 million cells was incubated with antibody-bound beads and then subjected to the ChIP-exo 5.0 assay (Rossi et al. 2018).
ChIP-exo 5.0 assay
Chromatin for ChIP-exo was prepared by resuspending cross-linked and quenched chromatin in Farnham cell lysis buffer at a ratio of 25 million cells to 1 mL of buffer for 20 min at 4°C. At the 10-min mark, cells were pushed through a 25-gauge needle five times to enhance cellular lysis. Nuclei were then isolated by pelleting at 2500g for 5 min. Nuclei were resuspended in RIPA buffer (25 million cells to 1 mL of buffer) for an additional 20 min at 4°C and then pelleted again at 2500g for 5 min. Disrupted nuclei were then finally resuspended in 1× PBS (25 million cells to 1 mL of buffer) and sonicated for 10 cycles (30-on/30-off) in a Diagenode pico. Solubilized chromatin was then processed through ChIP-exo. Production-scale ChIP-exo 5.0 was generally performed in batches of 48 in a 96-well plate, alternating every column to reduce risk of cross-contamination. Briefly, solubilized chromatin was incubated with pAG Dynabeads, preloaded with 3 ug of antibody overnight, then sequentially processed through A-tailing, first adapter ligation, Phi29 fill-in, lambda exonuclease digestion, cross-link reversal, second adapter ligation, and PCR for final high-throughput sequencing. Equal proportions of ChIP samples were barcoded, pooled, and sequenced. Illumina paired-end read (40-bp Read_1 and 36-bp Read_2) sequencing was performed on a NextSeq 500 and 550. Although, on average, we sought approximately 10 million total paired-end reads per ChIP, we accepted less if there was strong evidence of target enrichment. Otherwise, we performed an additional round of sequencing. The 5′ end of Read_1 corresponded to the exonuclease stop site, located ∼6 bp upstream of a protein–DNA cross-link. Read_2 served two indirect functions: to provide added specificity to genome-wide mapping and to remove PCR duplicates.
ChIP-seq
We fixed 1 × 108 cells in 1% formaldehyde (Sigma-Aldrich 252549) in PBS for 15–20 min at room temperature and quenched with 1/10th volume of 1.25 M glycine for 5 min at room temperature. Cells were collected at 1000g for 5 min, washed in PBS, and pelleted at 1000 × g for 5 min, and pellets were flash-frozen in liquid nitrogen and stored at −80°C until use. Pellets were thawed on ice and resuspended in 10 mL lysis buffer 1 (50 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% IGEPAL CA-630, 0.25% Triton X-100 with 5 µL/mL Sigma-Aldrich 8340 protease inhibitor cocktail), incubated on ice 10 min, pelleted 1500g, and subsequently washed in lysis buffer 2 (10 mM Tris-HCl at pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA and 5 uL/mL protease inhibitor) as with lysis buffer 1 before resuspending in 1 mL lysis buffer 3 (10 mM tris-HCl at pH 8.0, 1 mM EDTA, 0.1% SDS, and 5 uL/mL protease inhibitors) for sonication as previously described (Lee et al. 2006). Chromatin was sheared in a 1-mL milliTUBE with AFA fiber on a Covaris E220 using 10% duty factor for 2 min. The sheared chromatin concentration was estimated with NanoDrop at OD260 and diluted to 1 mg/mL in ChIP dilution buffer (10% Triton X-100, 1 M NaCl, and 1% sodium deoxycholate). One milligram chromatin was combined with 4 µg hybridoma tissue culture supernatant and rotated overnight at 4°C. Forty microliters of protein G Dynabeads was added and incubated for 2–4 h rotating at 4°C. Samples were washed five times with 1 mL RIPA buffer, two times with TE with 50 mM NaCl. Chromatin was eluted with 800 µL elution buffer (50 mM Tris at pH 8.0, 1 mM EDTA, 0.1% SDS) for 30 min at 65°C, shaking at 1500 rpm in a ThermoMixer (Eppendorf). Supernatants were collected, digested with 20 µL of 20 mg/mL Proteinase K, and incubated overnight at 65°C. DNA was purified with phenol chloroform extraction. Five hundred microliters of the aqueous phase was precipitated with 20 µL 5 M NaCl, 1.5 µg glycogen, and 1 mL EtOH for 1 h on ice or overnight at −20°C.
Sequencing libraries were prepared with the KAPA HTP library prep kit (Roche) using 1–10 ng DNA, and libraries were size-selected with AMPure XP beads (Beckman Coulter). Illumina 50-bp single-end read sequencing was performed on a NextSeq 500 or NovaSeq 6000. The modular pipeline Ceto (https://github.com/ebartom/NGSbartom) was used to convert base calls to FASTQ, align reads to BAM files, and make bigWig coverage tracks. Briefly, bcl2fastq with parameters -r 10 -d 10 -p 10 -w 10 was used to generate FASTQ files. Trimmomatic version 0.33 with the options single end mode (SE) and -phred33 was used to remove low-quality reads. Reads were then aligned to hg19 with Bowtie 1.1.2 (Langmead et al. 2009) with options -p 10 -m 1 -v 2 -S, thus keeping only uniquely mapped reads and allowing up to two mismatches. Coverage tracks were created with the R script from Ceto, createChIPtracks.R ‐‐extLen = 150 to extend reads to 150 bp, and coverage was normalized to total mapped reads (reads per million). Peaks were called with MACS2 2.1.0 (Zhang et al. 2008) with a cutoff of -q 0.01 and the input chromatin used as the control data set. Heatmaps and composite plots were made with deepTools (Ramírez et al. 2016) version 2.0. computeMatrix using reference-point peaks. “Blacklisted” regions were removed with parameter -bl Anshul_Hg19UltraHighSignalArtifactRegions.bed (ftp://encodeftp.cse.ucsc.edu/users/akundaje/rawdata/blacklists/hg19). For metagene plots, we used the Ensembl version 75 transcripts with the highest total coverage from the annotated TSS to 200 nt downstream and were at least 2 kb long, as well as 1 kb away from the nearest gene.
RNAi
Lentiviruses were packaged in HEK293T cells transfected with 1 µg pME-VSVG, 2 µg PAX2, and 4 µg shRNA in pLKO.1 backbone using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's instructions. The virus particles were then harvested 24–48 h later by passing through a 0.45-µm syringe filter (Thermo Fisher Scientific). Viruses were mixed with an equal volume of fresh media supplemented with 10% FBS, and polybrene was added at a final concentration of 5 µg/mL to increase infection efficiency. The medium was changed 6 h after infection. Cells were selected with 2 µg/mL puromycin for 3 d before western blotting (anti-NRF1 rabbit mAb clone D9K6R from CST and anti-tubulin beta mouse mAb clone E7 from DSHB) and ChIP-seq experiments. For the shRNA sequences used, see Table 1.
Table 1.
shRNA knockdown oligo sequences for NRF1
Bioinformatic protocols
Genome alignment
All genomic experiments generating sequencing reads were aligned to hg19. Because of the validation criteria selected, no results are expected to be different across human genome builds.
Technical performance
A series of modular bioinformatic analyses were implemented to evaluate technical success of ChIP-exo library construction and sequencing, independent of whether the antibody found its target or not: (1) sequencing depth (standard is 8–10 M), which is the total number of sequencing reads having a target-specific barcode; (2) adapter dimers (standard is <2%), which is the fraction of reads that contain the sequencing adapters but lack a genomic insert; (3) alignment (standard is 70%–90%), which is the percentage of reads that map to the reference human genome after removing adapter dimers; and (4) PCR duplicates (standard is <40%), which are expected to have identically mapped Read_1 and Read_2 5′ ends. We assume that when Read_1 and Read_2 5′ ends have identical mapped coordinates; they represent PCR duplicates. Because Read_2 is generated by sonication, it is expected to be distributed across a region and thus is not likely to be at the same coordinate twice. Because PCR duplicates are not a direct product of ChIP, they add no value to enrichment metrics. High PCR duplicates, at normal sequencing depths, often mean technical loss of material during library construction before PCR.
Peak calling
We used two distinct algorithms for ChIP-exo peak-calling. The first algorithm was GeneTrack, which used a Gaussian kernel to call strand-separated peaks at the 5′ ends of reads (Albert et al. 2008). The reads were then paired across strands, and the tag occupancy was normalized using the NCIS approach (Liang and Keleş 2012). Peak significance was called using either the binomial or Poisson test (taking whichever P-value was higher) with Benjamini–Hochberg correction and a Q-value cutoff of Q < 0.01. GeneTrack was used to generate the ChIP-exo peaks used in the manuscript figures. The other peak-caller used was the ChExMix algorithm, which is a high-resolution peak-caller designed to simultaneously identify enriched sequence motifs and distinct subtypes of binding using a combination of clustering and hierarchical mixture modeling (Yamada et al. 2019). ChExMix was designed to take advantage of ChIP-exo's ability to identify protein cofactors through indirect cross-linking events by modeling detected tag distributions in ChIP-exo data and using tree-clustering-based approaches to determine the significant peak subtypes that exist with ChIP-exo data. ChExMix-called peaks were used to interrogate the ChIP-exo peaks subtype structure and are visualized on the www.PCRPvalidation.org website.
Motif enrichment via ChExMix
De novo motif discovery was performed by ChExMix. Each motif was compared against the JASPAR database using TOMTOM with default parameters to identify similarity to known motifs (Bailey et al. 2009; Fornes et al. 2020). Heatmaps and composite plots were generated of sequence reads aligned relative to motif midpoints of all peaks containing an enriched motif. For samples with high background, low complexity, and/or low sequencing depth, it is possible that the antibody is valid but that standard de novo motif discovery may fail. We developed an orthogonal method for motif detection. By first identifying all nonredundant motif classes (Castro-Mondragon et al. 2017) in the genome, we then overlap low-threshold ChExMix peaks and determined which motif class possesses overlapping peaks above background (>2 log2).
Motif enrichment and centering analysis
For ChIP-exo data generated from K562, HepG2, and MCF-7 cells, narrowpeak data were called using ChExMix (Yamada et al. 2019); we restricted our motif enrichment analysis to narrowpeak data sets that contained more than 500 peaks. For ChIP-seq data generated from HCT116 cells, we required the presence of 100 peaks owing to the typical lower number of called peaks in those data sets compared with ChIP-exo. Motif enrichment analysis of ChIP-exo and ChIP-seq peaks was then performed as described previously (Mariani et al. 2017). For ChIP-exo peaks, we first filtered for the data sets that had more than 500 peaks and then used for the comparison the top 500 peaks, with peaks defined as the ChIP-exo summits computationally padded with the region spanning [−100 bp, +100 bp]. To perform an analogous analysis on ChIP-seq peaks, we fixed both the number of peaks per data set (e.g., top 100 peaks) and the peak size, which we computationally trimmed similarly to the ChIP-exo data to span [−100 bp, +100 bp] surrounding the ChIP-seq peak summit. For each ChIP peak set, we generated background sequences using GENRE software with the default human setting to ensure the same level of promoter overlap, repeat overlap, GC content, and CpG dinucleotide frequency between each peak and its associated background sequence (Mariani et al. 2017). We manually curated a collection of 100 PWMs, primarily from biochemical TF DNA-binding assays (i.e., PBM or HT-SELEX), from the UniPROBE and Cis-BP databases, as a representative repertoire of human sequence-specific TF binding motifs (Weirauch et al. 2014; Hume et al. 2015; Mariani et al. 2020). We scored each sequence for matches to each of the motifs using the function “matchPWM” from the “Biostrings” R package (R Core Team 2020). Motif enrichment was quantified using an established AUROC metric that assesses the presence of a motif among the 500 highest-confidence peaks (foreground set) compared with the corresponding background set of sequences using publicly available tools for analysis of TF ChIP-seq data (Mariani et al. 2017). We also assessed each motif for its enrichment toward the centers of each ChIP-exo or ChIP-seq peak set as described previously (Mariani et al. 2017). Briefly, we first identified the PWM score threshold that maximized the difference between foreground and corresponding background sets in the number of sequences containing at least one PWM match (optimal PWM match score). If a sequence had multiple PWM matches, we considered only the highest score site. We then calculated the distance from each of these sites to the corresponding peak summits and used the mean of these distances in the foreground or background set to quantify the motif enrichment toward the centers of ChIP peaks. The P-values associated with motif enrichment (i.e., AUROC value) and enrichment toward the peak summits (i.e., mean motif distance to peak summit) were both calculated by using a Wilcoxon signed-rank test comparing their scores (PWM match score and PWM match distance to distance to peak summit, respectively) for foreground and background sequences when the PWM threshold was set to the optimal PWM match score. We then adjusted the P-values across the PWM collection with a false-discovery rate test for multiple hypothesis testing. To test the significance of the difference in the TPM distributions between ChIP data sets with enriched versus nonenriched motifs, we calculated the P-value by a Wilcoxon test using the function wilcox.test in R (R Core Team 2020).
Genome annotation enrichment
Only a small fraction of DNA-interacting factors binds sequence-specific motifs. In the case of targets with either no expected motif or no known function, determining peak enrichments at annotated regions of the genome can provide evidence of ChIP success. The relative frequency of peaks occurring in different functional genomic regions as defined by ChromHMM was calculated for each target, an IgG-negative control, and random expectation (Ernst et al. 2011). The log2 frequency enrichment of sample over IgG control was used to identify regions of enrichment, as well as significant areas of de-enrichment (regions that selectively avoid the target). Significant peaks were intersected with ChromHMM and Segway states to generate frequency histograms for overlap with predicted chromatin states (Ernst et al. 2011; Hoffman et al. 2012). Peaks derived from the matched negative control data set were also intersected with annotated states. The log2 ratio of sample state frequency over control state frequency was then calculated to identify general state enrichment of the sample throughout the genome.
Positional enrichment at promoters and insulators
To identify enrichment in well-characterized promoter regions, sequence reads for the target, a matched “no antibody” control, and an IgG were aligned relative to annotated TSSs. Heatmaps of all genes and composite plots of the top 1000 TSSs by gene expression (RNA-seq FPKM) were generated from the data.
Heatmaps, composite plots, and four-color sequence plots
All heatmaps, composite plots, and four-color sequence plots were generated using ScriptManager v0.12 (https://github.com/CEGRcode/scriptmanager). ScriptManager is a Java-based GUI tool that contains a series of interactive wizards that guide the user through transforming aligned BAM files into publication-ready figures.
CUT&RUN protocols
Antibody sourcing and concentration
Antibody hybridoma supernatants, name, clone ID, and lot) were from DSHB. mAbs were concentrated using Amicon ultra-4 centrifugal filter units with a 50-kDa cut-off (MilliporeSigma UFC805024) following manufacturer's recommendations. All centrifugation steps (including 3 × 4 mL washes with 1× Tris buffered Saline [TBS]) were performed at 4000 g for 15 min at room temperature. Final concentrations for recovered mAbs (stored at 4°C in TBS, 0.1% BSA, 0.09% sodium azide) were assumed based on initial concentrations/final recovery volumes and 1 µg used per CUT&RUN experiment.
CUT&RUN
CUT&RUN was performed on 500,000 native nuclei extracted from K562 cells using CUTANA protocol v1.5.1 (http://www.epicypher.com), which is an optimized version of that previously described (Skene et al. 2018). For each sample, nuclei were extracted by incubating cells on ice for 10 min in nuclei extraction buffer (NE: 20 mM HEPES–KOH at pH 7.9; 10 mM KCl; 0.1% Triton X-100; 20% glycerol; 0.5 mM spermidine; 1× complete protease inhibitor; Roche 11836170001), collecting by centrifugation (600g, 3 min, 4°C), discarding the supernatant, and resuspending at [100 µL/500 K nuclei] sample in NE buffer. For each target, 500,000 nuclei were immobilized onto Concanavalin-A beads (EpiCypher #21-1401) and incubated overnight (4°C with gentle rocking) with 1 µg of antibody (for all 40 PCRP antibodies as above; RbIgG [EpiCypher 13-0042, lot 20036001-52]; MsIgG [Invitrogen 10400C, lot VD293456]; CTCF [Millipore 07-729, lot 3205452]).
Modified CUT&RUN library prep
Illumina sequencing libraries were prepared from 1 ng to 10 ng of purified CUT&RUN DNA using NEBNext ultra II DNA library prep kit (New England Biolabs E7645) as previously described (Liu et al. 2018) with the following modifications to preserve and enrich smaller DNA fragments (20–70 bp). Briefly, during end repair, the cycling time was decreased to 30 min at 50°C. After adapter ligation, to purify fragments >50 bp, 1.75× volumes of Agencourt AMPure XP beads (Beckman Coulter A63881) were added for the first bead clean-up before amplification following manufacturer's recommendations. PCR amplification cycling parameters were as previously described (Skene et al. 2018). Post PCR, two rounds of DNA size-selection were performed. For the first selection, 0.8× volume of AMPure XP beads was added to the PCR reaction to remove products >350 bp. The supernatant, containing fragments <350 bp, was moved forward to a second round of size-selection using 1.2× volumes of AMPure XP beads to remove products <150 bp. Libraries were quantified using a Qubit fluorometer (Invitrogen) and checked for size distribution with a Bioanalyzer (Agilent).
CUT&RUN library sequencing and data analysis
Libraries were sequenced on the Illumina NextSeq 550, obtaining approximately 5 million paired-end reads (75 × 75 nucleotides) on average. Paired-end FASTQ files were aligned to the hg19 reference genome using the ChIP-exo pipeline.
TF cloning, protein expression, western blots, and PBM protocols
Full-length TFs were either obtained from the hORFeome clone collection or synthesized as gBlocks (Integrated DNA Technologies) (Supplemental Table 8), full-length sequence-verified, and transferred by Gateway recombinational cloning into either the pDEST15 (Thermo Fisher Scientific) or pT7CFE1-NHIS-GST (Thermo Fisher Scientific) vectors for expression as N-terminal GST fusion proteins (ORFeome Collaboration 2016). TFs were expressed by a coupled IVT kit according to the manufacturer's protocols (Supplemental Table 7). Protein concentrations were approximated by an anti-GST western blot as described previously (Berger et al. 2006). All PCRP antibodies were used at a final concentration of 40 ng/mL in western blots; based on successful outcomes in PBM experiments, PCRP antibodies 1A7 (anti-GATA4), 2A4 (anti-HNF4A), and 1B3 (anti-PAX3) were also used at a final concentration of 1000 ng/mL in western blots. The 8 × 60 K GSE “all 10-mer universal” oligonucleotide arrays (AMADID 030236; Agilent Technologies) were double-stranded and used in PBM experiments essentially as described previously, with minor modifications as described below (Berger et al. 2006; Berger and Bulyk 2009; Nakagawa et al. 2013). GST-tagged TFs assayed in PBMs were detected either with Alexa Fluor 488–conjugated anti-GST antibody (Invitrogen A-11131), or with a TF-specific PCRP antibody, followed by washes and detection with Alexa Fluor 488–conjugated goat antimouse IgG(H + L) cross-adsorbed secondary antibody (Invitrogen A-11001), essentially as described previously (Supplemental Table 7; Siggers et al. 2011b). All PCRP Abs were used undiluted in PBM experiments; a subset of the PCRP Abs were also tested at a 1:5 or 1:20 dilution (Supplemental Table 7). All PBM experiments using PCRP antibodies were performed using fresh arrays or arrays that had been stripped once, as described previously (Berger et al. 2006; Berger and Bulyk 2009). PBMs were scanned in a GenePix 4400A microarray scanner, and raw data files were quantified and processed using the Universal PBM Analysis Suite (Berger et al. 2006; Berger and Bulyk 2009).
STORM protocols
Supernatant concentration
3 milliliters of PCRP supernatant were concentrated using the Amicon pro affinity concentration kit Protein G with a 10-kDa Amicon ultra-0.5 device following the manufacturer's recommendations. Supernatant was quantitated by spectrophotometer for rough approximation of concentration.
Cellular preparation and staining
K562 cells obtained from the ATCC (CCL243) were grown in a humidified 5% CO2 incubator; 3–5 × 105 cells were centrifuged at 1500 rpm for 5 min, washed with PBS, and plated on MatTek-brand glass-bottom dishes (P35-G.1) prepared appropriately (washes with increasing concentrations of ethanol, followed by coating with poly-L-Lysine [Sigma-Aldrich P4707] for 5 min and subsequent washes with water and airdrying for 2 h) After plating, the cells were allowed to adhere for 2 h and then washed with PBS. For fixation, 1 mL of 4% paraformaldehyde and 0.1% glutaraldehyde in PBS was added for 10 min at room temperature with gentle rocking followed by blocking and permeabilization in 2% normal goat serum/1% Triton X-100 in PBS for 1 h at room temperature with gentle shaking. Immunostaining was performed with anti-MTR4 antibody (Abcam 70551) at a dilution of 1:250 in 0.1% normal goat serum, 0.05% Triton X-100 overnight at 4°C. The next morning, cells were incubated with the secondary antibody (conjugated to Alexa Fluor 647; Invitrogen A31571) at a 1:1000 dilution in 0.1% normal goat serum in PBS and incubated for 2 h at room temperature followed by washes in PBS. This material was then subjected to immunostaining with PCRP antibodies by repeating the above procedure. PCRP mAb hybridoma supernatant (3 mL) was first concentrated 30- to 100-fold because raw supernatants were unsuccessful in both confocal microscopy and STORM. The secondary antibody was conjugated to Alexa Fluor 488 (Invitrogen A21206). At the end of the application of the second secondary antibody, the cells were washed three times with PBS. Subsequently, DAPI (Sigma-Aldrich D8542) at a concentration of 1:500 in 0.1% normal goat serum in 1× PBS was applied for 10 min at room temperature. The cells were washed three times with PBS, and finally, 4% paraformaldehyde/0.1% glutaraldehyde in PBS was applied for 10 min at RT before three washes in 1× PBS were performed and dishes stored at 4°C until microscopy could be performed.
Microscopy
Cells were brought to the imaging facility, and OXEA buffer was applied (50 mM cysteimine, 3% v/v oxyfluor, 20% v/v sodium DL lactate, with pH adjusted to approximately 8.5, as necessary). The two colors (Alexa Fluor 488 and Alexa Fluor 647) were imaged sequentially. Imaging buffer helped to keep dye molecules in a transient dark state. Subsequently, individual dye molecules were excited stochastically with high laser power at their excitation wavelength (488 nm for Alexa Fluor 488 or 647 nm for Alexa Fluor 647, respectively) to induce blinking on millisecond timescales. STORM images and the correlated high-power confocal stacks were acquired via a CFI Apo TIRF 100 × objective (1.49 NA) on a Nikon Ti-E inverted microscope equipped with a Nikon N-STORM system, an Agilent laser launch system, an Andor iXon ultra 897 EMCCD (with a cylindrical lens for astigmatic 3D-STORM imaging) camera, and an NSTORM Quad cube (Chroma). This setup was controlled by Nikon NIS-Element AR software with N-STORM module. To obtain images, the field of view was selected based on the live EMCCD image under 488-nm illumination. 3D STORM data sets of 50,000 frames were collected. Lateral drift between frames was corrected by tracking 488, 561, and 647 fluorescent beads (TetraSpeck, Invitrogen). STORM images were processed to acquire coordinates of localization points using the N-STORM module in NIS-Elements AR software. Identical settings were used for every image. Each localization is depicted in the STORM image as a Gaussian peak, the width of which is determined by the number of photons detected (Betzig et al. 2006). All of the 3D STORM imaging was performed on a minimum of two different K562 cells.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE151287, GSE152144, and GSE151326. PBM data have been deposited in the UniPROBE database (http://the_brain.bwh.harvard.edu/uniprobe/downloads.php under Lai et al. 2020) (Hume et al. 2015). Code to run the ChIP-exo analysis pipeline is available as Supplemental Code and at GitHub (https://github.com/CEGRcode/PCRPpipeline). Peak files for all figures are available as Supplemental Data and at GitHub (https://github.com/CEGRcode/2021-Lai_PCRP).
Supplementary Material
Acknowledgments
The Basu laboratory thanks Drs. Teresa Swayne and Emilia Laura Munteanu of the Herbert Irving Comprehensive Cancer Center Core Microscopy facility at Columbia University. The Pugh laboratory thanks Kyle Nilson for donating the peroxide-stressed cells. The Shilatifard laboratory thanks Anna Whelan and Jordan Harris for technical assistance and Marc Morgan for helpful discussions. The Bulyk laboratory thanks Steve S. Gisselbrecht for assistance with preparation of figures. ChIP-exo data were made available through the Cornell Institute of Biotechnology's Epigenomic Core Facility using the Platform for Epigenome and Genomic Research (PEGR), with National Institutes of Health (NIH) 5R01-ES013768-12 funding for the development and dissemination of PEGR. We acknowledge the support of the Institute for Computational and Data Sciences at the Pennsylvania State University through an ICDS Seed Grant. This work was supported by an administrative supplement to National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI099195 to U.B., an administrative supplement to NIH grant R21 HG009268 to M.L.B., NIH grant R01-GM125722 and an administrative supplement to NIH grant R01-ES013768 to B.F.P., an administrative supplement to NIH grant R01CA214035 to A.S., NIH grant R50CA211428 to E.R.S., and NIH grant R44 DE029633 to EpiCypher.
Author contributions: In the Basu laboratory, G.R. performed STORM experiments; W.Z. performed required bioinformatics analyses of STORM data; and U.B. analyzed data, provided oversight, and cowrote the manuscript. In the Bulyk laboratory, J.T.A. and S.K.P. performed cloning, protein expression, western blots, PBM experiments, and PBM analysis; L.M. performed analysis of motif enrichment and centering; S.K.P. and L.M. prepared figures and supplemental tables; M.L.B. supervised research; and S.K.P., L.M., and M.L.B. cowrote the manuscript. In the Pugh laboratory, T.R.B., K.B., J.M., S.N.D., and K.M. performed ChIP-exo assays. P.K.K. designed and implemented the web portal. M.J.R. and D.J. performed ChIP-exo library quantitation and sequencing. W.K.M.L. directed the ChIP-exo experiments, processed and analyzed the ChIP-exo, and cowrote the manuscript writing. B.F.P. provided oversight for ChIP-exo and cowrote the manuscript. In the Shilatifard laboratory, A.P.S. and Z.Z. conducted experiments. E.R.S. analyzed ChIP-seq data. E.R.S. and A.S. provided oversight and cowrote the manuscript. From EpiCypher, B.J.V., K.N., and E.M. performed CUT&RUN studies, and M-C.K. provided oversight and cowrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275472.121.
Competing interest statement
M.L.B. is a coinventor on U.S. patent 8,530,638 on universal PBM technology. B.F.P. has a financial interest in Peconic, LLC, which uses the ChIP-exo technology implemented in this study and could potentially benefit from the outcomes of this research. EpiCypher is a commercial developer of reagents to support CUTANA CUT&RUN. The authors in the Basu and Shilatifard laboratories declare no competing financial interests.
References
- Albert I, Wachi S, Jiang C, Pugh BF. 2008. GeneTrack—a genomic data processing and visualization framework. Bioinformatics 24: 1305–1306. 10.1093/bioinformatics/btn119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Machanick P. 2012. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128. 10.1093/nar/gks433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. 2015. Reproducibility crisis: blame it on the antibodies. Nature 521: 274–276. 10.1038/521274a [DOI] [PubMed] [Google Scholar]
- Baler R, Dahl G, Voellmy R. 1993. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol Cell Biol 13: 2486–2496. 10.1128/mcb.13.4.2486-2496.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Bulyk ML. 2009. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat Protoc 4: 393–411. 10.1038/nprot.2008.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW 3rd, Bulyk ML. 2006. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24: 1429–1435. 10.1038/nbt1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313: 1642–1645. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Venkataraman A, Irizarry J, Yang K, Anderson S, Campbell E, Gatlin CL, Freeman NL, Basavappa R, Stewart R, et al. 2016. The NIH Protein Capture Reagents Program (PCRP): a standardized protein affinity reagent toolbox. Nat Methods 13: 805–806. 10.1038/nmeth.4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogani D, Morgan MA, Nelson AC, Costello I, McGouran JF, Kessler BM, Robertson EJ, Bikoff EK. 2013. The PR/SET domain zinc finger protein Prdm4 regulates gene expression in embryonic stem cells but plays a nonessential role in the developing mouse embryo. Mol Cell Biol 33: 3936–3950. 10.1128/MCB.00498-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Mondragon JA, Jaeger S, Thieffry D, Thomas-Chollier M, van Helden J. 2017. RSAT matrix-clustering: dynamic exploration and redundancy reduction of transcription factor binding motif collections. Nucleic Acids Res 45: e119. 10.1093/nar/gkx314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chames P, Van Regenmortel M, Weiss E, Baty D. 2009. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol 157: 220–233. 10.1111/j.1476-5381.2009.00190.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Renewable Protein Binder Working Group, Gräslund S. 2011. A roadmap to generate renewable protein binders to the human proteome. Nat Methods 8: 551–558. 10.1038/nmeth.1607 [DOI] [PubMed] [Google Scholar]
- Edfors F, Hober A, Linderbäck K, Maddalo G, Azimi A, Sivertsson A, Tegel H, Hober S, Szigyarto CA, Fagerberg L, et al. 2018. Enhanced validation of antibodies for research applications. Nat Commun 9: 4130. 10.1038/s41467-018-06642-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. 2011. An assessment of histone-modification antibody quality. Nat Struct Mol Biol 18: 91–93. 10.1038/nsmb.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen E, Brandsma JH, Moen MJ, Signorile L, Dekkers DH, Demmers J, Kockx CE, Ozgür Z, van IJcken WF, van den Berg DL, et al. 2015. Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat Commun 6: 7155. 10.1038/ncomms8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranasic D, et al. 2020. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 48: D87–D92. 10.1093/nar/gkz1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan R, Hartemink AJ, Bulyk ML. 2009. Distinguishing direct versus indirect transcription factor–DNA interactions. Genome Res 19: 2090–2100. 10.1101/gr.094144.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. 2007. Quantifying similarity between motifs. Genome Biol 8: R24. 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanly WC, Artwohl JE, Bennett BT. 1995. Review of polyclonal antibody production procedures in mammals and poultry. ILAR J 37: 93–118. 10.1093/ilar.37.3.93 [DOI] [PubMed] [Google Scholar]
- Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. 2012. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods 9: 473–476. 10.1038/nmeth.1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E, et al. 2013. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res 41: 827–841. 10.1093/nar/gks1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsby M, Paduch M, Miersch S, Sääf A, Matsuguchi T, Lee B, Wypisniak K, Doak A, King D, Usatyuk S, et al. 2015. A high through-put platform for recombinant antibodies to folded proteins. Mol Cell Proteomics 14: 2833–2847. 10.1074/mcp.O115.052209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. 2015. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein–DNA interactions. Nucleic Acids Res 43: D117–D122. 10.1093/nar/gku1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G, Milstein C. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256: 495–497. 10.1038/256495a0 [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831. 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1: 729–748. 10.1038/nprot.2006.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Keleş S. 2012. Normalization of ChIP-seq data with control. BMC Bioinformatics 13: 199. 10.1186/1471-2105-13-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Giri PK, Kazadi D, Laffleur B, Zhang W, Grinstein V, Pefanis E, Brown LM, Ladewig E, Martin O, et al. 2017. Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell 169: 523–537.e15. 10.1016/j.cell.2017.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Fallahi-Sichani M, Sorger PK. 2015. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun 6: 8390. 10.1038/ncomms9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK. 2014. The history of monoclonal antibody development – progress, remaining challenges and future innovations. Ann Med Surg (Lond) 3: 113–116. 10.1016/j.amsu.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Hargreaves VV, Zhu Q, Kurland JV, Hong J, Kim W, Sher F, Macias-Trevino C, Rogers JM, Kurita R, et al. 2018. Direct promoter repression by BCL11A controls the fetal to adult hemoglobin switch. Cell 173: 430–442.e17. 10.1016/j.cell.2018.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood T, Yang PC. 2012. Western blot: technique, theory, and trouble shooting. N Am J Med Sci 4: 429–434. 10.4103/1947-2714.100998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani L, Weinand K, Vedenko A, Barrera LA, Bulyk ML. 2017. Identification of human lineage-specific transcriptional coregulators enabled by a glossary of binding modules and tunable genomic backgrounds. Cell Syst 5: 187–201.e7. 10.1016/j.cels.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani L, Weinand K, Gisselbrecht SS, Bulyk ML. 2020. MEDEA: analysis of transcription factor binding motifs in accessible chromatin. Genome Res 30: 736–748. 10.1101/gr.260877.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. 2019. What to do about those immunoprecipitation blues. Nat Methods 16: 289–292. 10.1038/s41592-019-0365-3 [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. 2004. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet 36: 1331–1339. 10.1038/ng1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. 2013. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc Natl Acad Sci 110: 12349–12354. 10.1073/pnas.1310430110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ORFeome Collaboration. 2016. The ORFeome Collaboration: a genome-scale human ORF-clone resource. Nat Methods 13: 191–192. 10.1038/nmeth.3776 [DOI] [PubMed] [Google Scholar]
- Park PJ. 2009. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 10: 669–680. 10.1038/nrg2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, Komorowski J, Gingeras TR, Wadelius C. 2008. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res 18: 380–392. 10.1101/gr.6880908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Reardon S. 2016. Thousands of goats and rabbits vanish from major biotech lab. Nature 10.1038/nature.2016.19411 [DOI] [Google Scholar]
- Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147: 1408–1419. 10.1016/j.cell.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2012. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr Protoc Mol Biol Chapter 21: Unit 21 24. 10.1002/0471142727.mb2124s100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. 2009. The role of DNA shape in protein–DNA recognition. Nature 461: 1248–1253. 10.1038/nature08473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi MJ, Lai WKM, Pugh BF. 2018. Simplified ChIP-exo assays. Nat Commun 9: 2842. 10.1038/s41467-018-05265-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah RN, Grzybowski AT, Cornett EM, Johnstone AL, Dickson BM, Boone BA, Cheek MA, Cowles MW, Maryanski D, Meiners MJ, et al. 2018. Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol Cell 72: 162–177.e7. 10.1016/j.molcel.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. 2011a. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat Immunol 13: 95–102. 10.1038/ni.2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers T, Duyzend MH, Reddy J, Khan S, Bulyk ML. 2011b. Non-DNA-binding cofactors enhance DNA-binding specificity of a transcriptional regulatory complex. Mol Syst Biol 7: 555. 10.1038/msb.2011.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski K, Mehta A, Inngjerdingen M, Thakor F, Kling S, Kalina T, Nyman TA, Stensland ME, Zhou W, de Souza GA, et al. 2018. A high-throughput pipeline for validation of antibodies. Nat Methods 15: 909–912. 10.1038/s41592-018-0179-8 [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, Henikoff S. 2018. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13: 1006–1019. 10.1038/nprot.2018.015 [DOI] [PubMed] [Google Scholar]
- Son KK, Tkach D, Rosenblatt J. 2001. Delipidated serum abolishes the inhibitory effect of serum on in vitro liposome-mediated transfection. Biochim Biophys Acta 1511: 201–205. 10.1016/S0005-2736(01)00297-8 [DOI] [PubMed] [Google Scholar]
- Trescher S, Leser U. 2019. Estimation of transcription factor activity in knockdown studies. Sci Rep 9: 9593. 10.1038/s41598-019-46053-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Björling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, et al. 2005. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4: 1920–1932. 10.1074/mcp.M500279-MCP200 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, Rimm DL, Rodriguez H, Hiltke T, Snyder M, et al. 2016. A proposal for validation of antibodies. Nat Methods 13: 823–827. 10.1038/nmeth.3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman A, Yang K, Irizarry J, Mackiewicz M, Mita P, Kuang Z, Xue L, Ghosh D, Liu S, Ramos P, et al. 2018. A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors. Nat Methods 15: 330–338. 10.1038/nmeth.4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22: 1798–1812. 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle FC, Tan H. 2015. A ChIP on the shoulder? Chromatin immunoprecipitation and validation strategies for ChIP antibodies. F1000Res 4: 235. 10.12688/f1000research.6719.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. 10.1016/j.cell.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. 1994. Making antibodies by phage display technology. Annu Rev Immunol 12: 433–455. 10.1146/annurev.iy.12.040194.002245 [DOI] [PubMed] [Google Scholar]
- Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. 2005. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res 33: W389–W392. 10.1093/nar/gki439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N, Lai WKM, Farrell N, Pugh BF, Mahony S. 2019. Characterizing protein–DNA binding event subtypes in ChIP-exo data. Bioinformatics 35: 903–913. 10.1093/bioinformatics/bty703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.