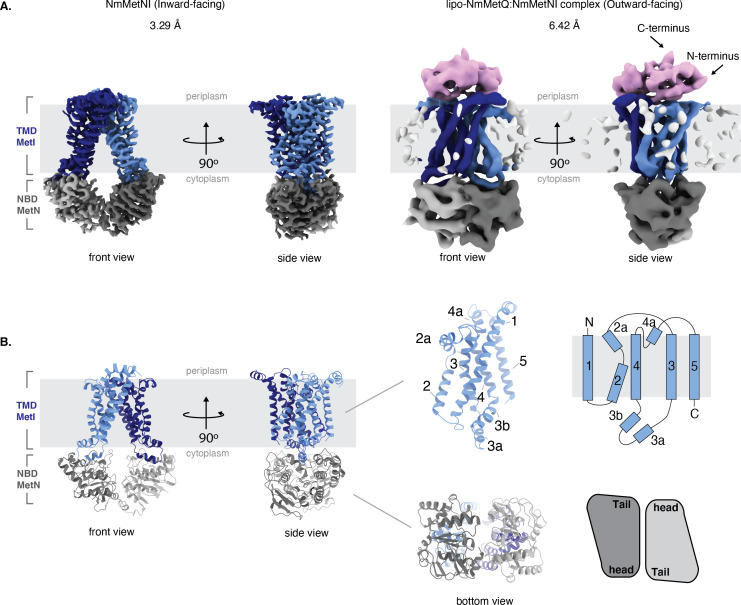
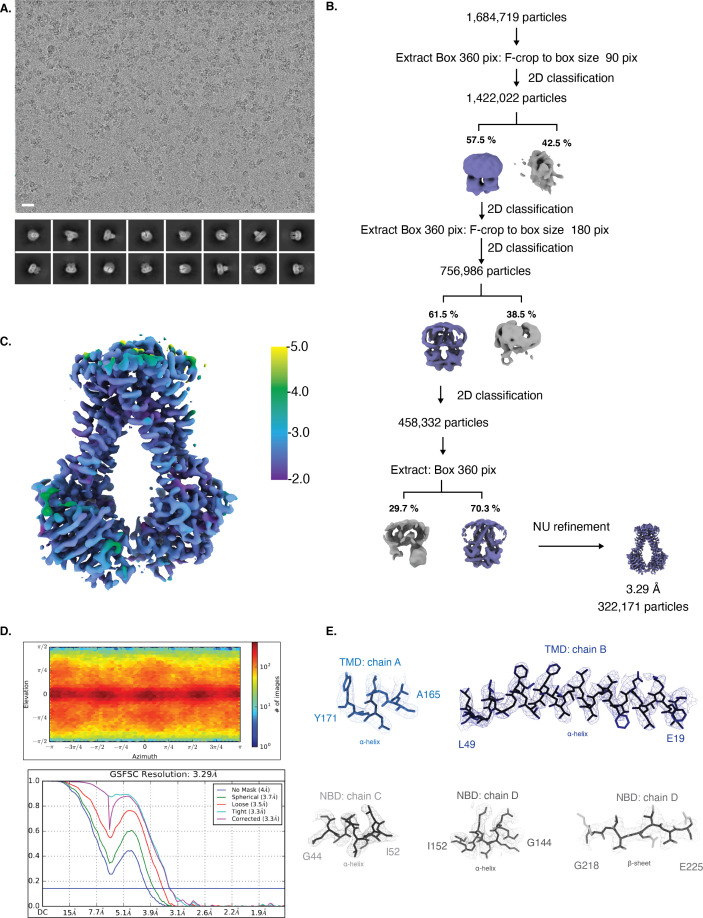
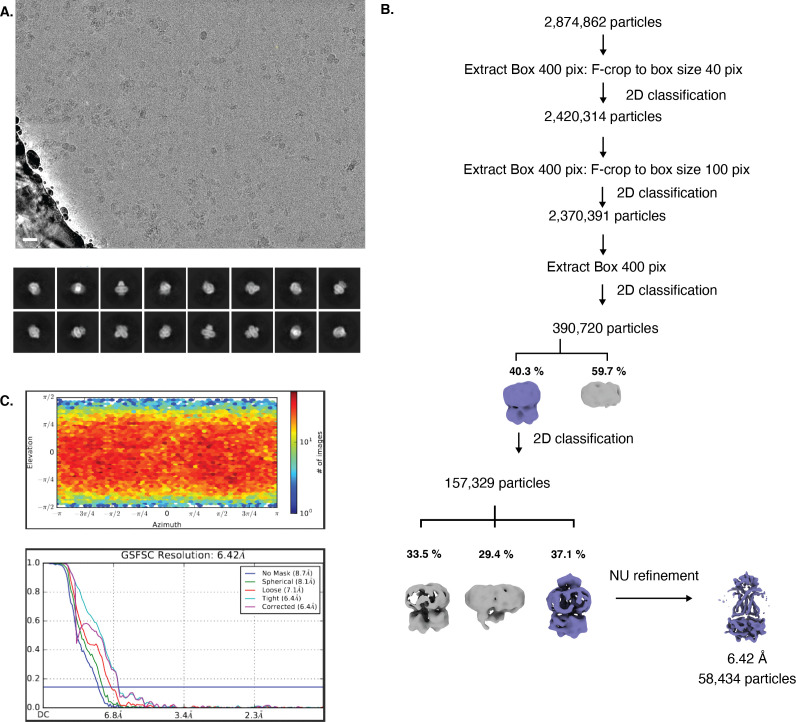
Figure 4. Architecture of NmMetNI and lipo-NmMetQ:NmMetNI complex.
(A) The 3.3 Å resolution cryo-EM map and NmMetNI in the inward-facing conformation in two views. (B) Transmembrane localization of NmMetI, showing NmMetI contains five transmembrane helices per monomer (C) The 6.4 Å resolution cryo-EM map and model of NmMetNI in complex with lipo-NmMetQ in the presence of ATP. NmMetNI is in the outward-facing conformation. NmMetI is shown in light/dark blue, NmMetN in light/dark grey and lipo-NmMetQ in light pink. The membrane is represented by a gray box.