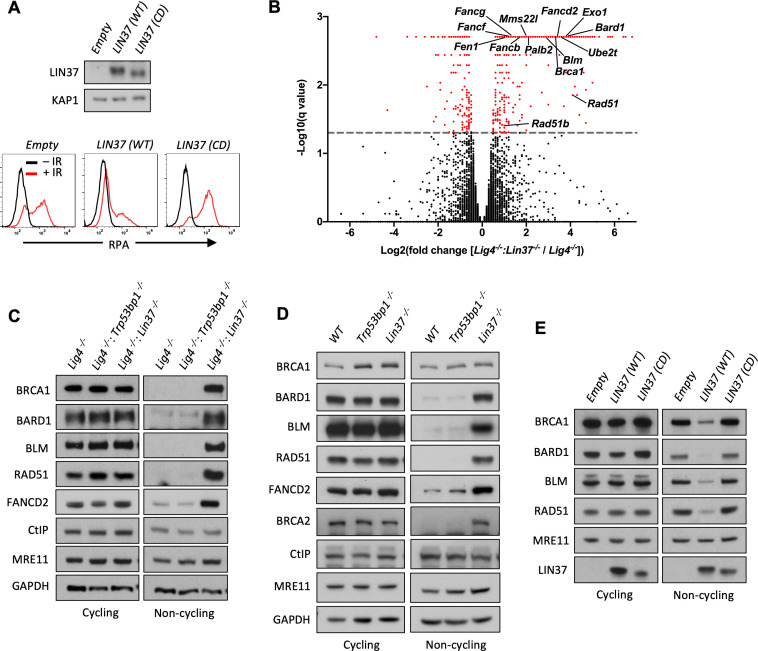
Figure 5. LIN37 suppresses the expression of HR protein expression in non-cycling cells.
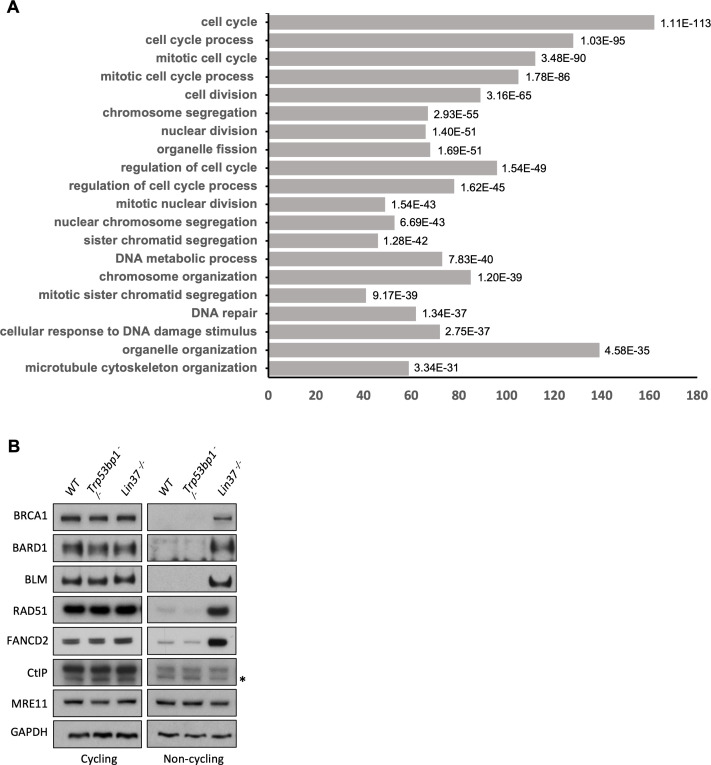
(A) Western blot analysis (top) and flow cytometric analysis for chromatin-bound RPA after before or after IR (bottom) of non-cycling Lig4−/−:Lin37−/− abl pre-B cells with empty lentivirus or lentivirus expressing wild type (WT) LIN37 or the LIN37 (CD) mutant. Representative of three experiments. (B) Volcano plot of RNA-Seq analysis of non-cycling Lig4−/− and Lig4−/−:Lin37−/− abl pre-B cells showing log2 values of the ratio of normalized transcript levels of Lig4−/−:Lin37−/− to Lig4−/− cells (X-axis) and −log10 of the q-values of fold enrichment of each gene (Y-axis). The dashed line indicates q=0.05. Genes with q-values≤0.05 are denoted as red dots. (C) Western blot analysis of indicated proteins in cycling and non-cycling Lig4−/−, Lig4−/−:Trp53bp1−/−, and Lig4−/−:Lin37−/− abl pre-B cells. (D) Western blot analysis of indicated proteins in cycling or non-cycling WT, Trp53bp1−/−, and Lin37−/− MCF10A cells. (E) Western blot analysis of indicated proteins in cycling and non-cycling Lig4−/−:Lin37−/− abl pre-B cells with empty lentivirus or lentivirus expressing WT LIN37 or the LIN37 (CD) mutant. HR, homologous recombination; IR, ionizing radiation; RNA-Seq, RNA sequencing.