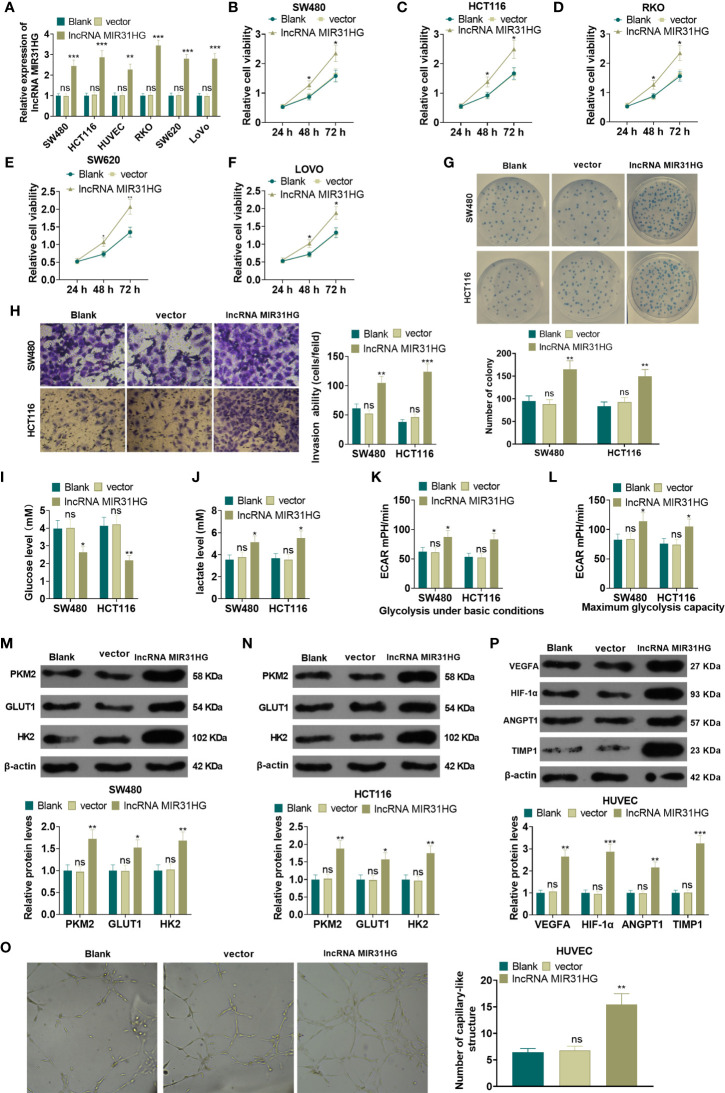
Figure 2.
Overexpressing MIR31HG facilitated CRC cell proliferation and glycolysis and endothelial cell angiogenesis. The MIR31HG overexpression model was constructed in CRC cell lines (Caco-2, RKO, SW480, SW620, LoVo and HCT116) and HUVEC cell lines. (A) The transfection validity was examined by qRT-PCR. (B–F) MTT assay was used to detect the proliferation of Caco-2, RKO, SW480, SW620, LoVo and HCT116 cells. (G) CRC cell proliferation was tested by colony formation assay. (H) Transwell assay were employed to evaluate CRC cell invasion. (I, J) The glucose and lactic acid production of CRC cells were measured using a glucose detection kit and lactic acid detection kit. (K, L) Glycolytic stress test was utilized to monitor the glycolytic level of CRC cells. (M, N) The expression of glycolysis-related proteins (PKM2, GLUT1, and HK2) in CRC cells was compared by WB. (O) Tube formation assay was used for evaluating the angiogenesis of HUVECs in vitro. The number of capillary-like structures was counted. (P) Western blot was used for detecting the expression of angiogenesis markers (including VEGFA, ANGPT1, HIF-1α and TIMP1) in HUVECs with MIR31HG overexpression. NS p > 0.05 (vs. Blank group), *P < 0.05, **P < 0.01, ***P < 0.01 (vs. vector group). (n=3).