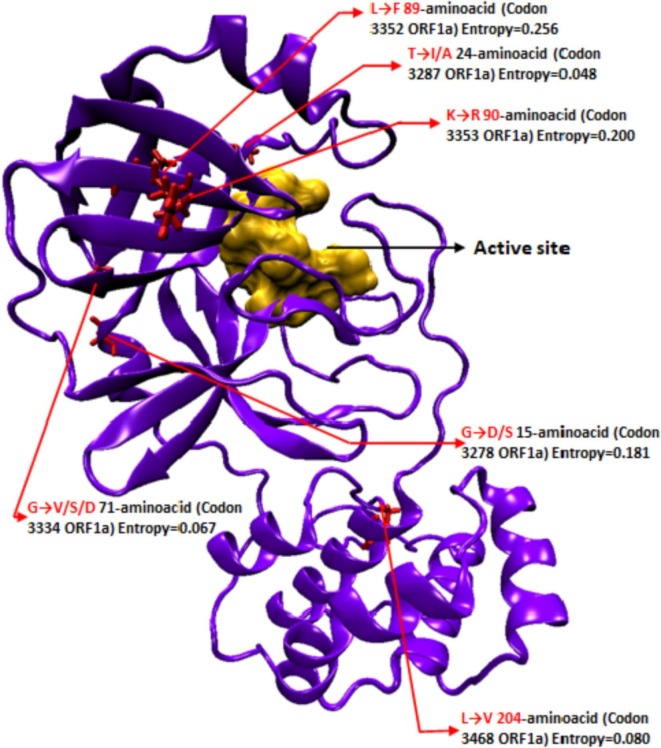
Fig. 1.
Schematic representation of relative locations of mutations in 3CLpro w.r.t the active site; Orange, active site surface model; red, mutant sites; Purple, protein backbone. Active site amino acid residues i.e. 39, 41, 145, 163, 164, and 165. Mutations are labeled with Amino acid substitution, amino acid position in 3CLpro peptide, and codon position in parent ORF1a with current global mutational frequency in form of entropy (GSAID). Mutations with very low frequency (Entropy < 0.05; 3955 genomes sequenced) were not included. Note: None of the mutations are in proximity of the active site/substrate binding region used as the receptor grid zone for the virtual screening of the compound libraries.