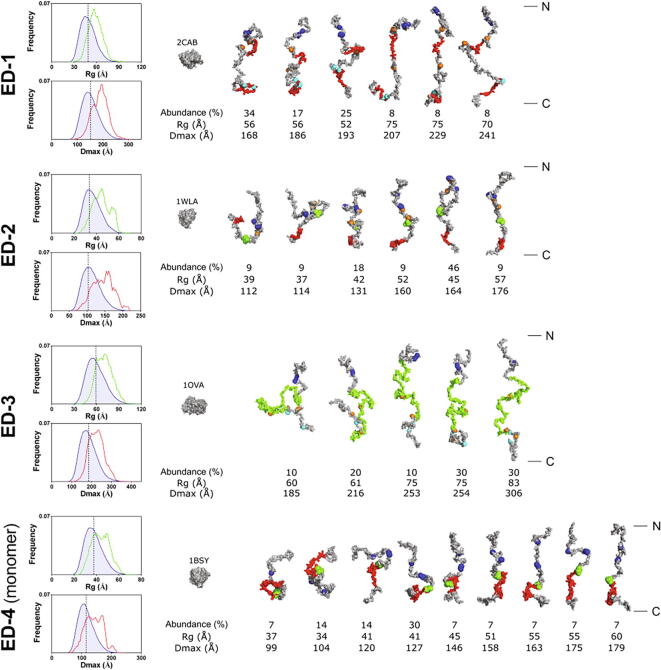
Fig. 4.
Ensemble Optimization Method analysis of SAXS data. The Rg and Dmax distributions derived from EOM analysis are shown on the left. The blue line shows the distribution of the initial pool of random coil conformers, whereas the green and red lines represent respectively the Rg and Dmax distributions of the selected conformer ensembles fitting the SAXS data. The dotted vertical black line represents the average Rg and Dmax of the random coil initial pool. The models issued from EOM deconvolution carried out with the parameters generated for RC proteins are represented with their relative abundance (%), Rg and Dmax. Backbone and side chains of EOM conformers were reconstructed to visualize biologically relevant sequences or amino acid residues. Serine residues bearing heparan sulfate or chondroitin sulfate chains in the proteoglycan form of syndecans are blue and cyan spheres respectively. The synstatin sequences of ED-1 (residues 93–120 and 210–240) and ED-4 (residues 87–131) are in red. The binding site of CD148 on ED-2 (residues 123–140) is in red and the NXIP motif of ED-2 (residues 95–98) and ED-4 (residues 87–90) is represented as green spheres. The mucin-like sequence of ED-3 is in green (residues 115–302), whereas the residues corresponding to single nucleotide polymorphisms are represented as orange spheres. Globular proteins with SASA values similar to those of the most compact conformations of the EDs are drawn to scale with their PDB code (PDB 2CAB: carbonic anhydrase, 1WLA: myoglobin, 1OVA: ovalbumin, 1BSY: β lactoglobulin). N- and C-termini of the EDs conformers are indicated on the right.