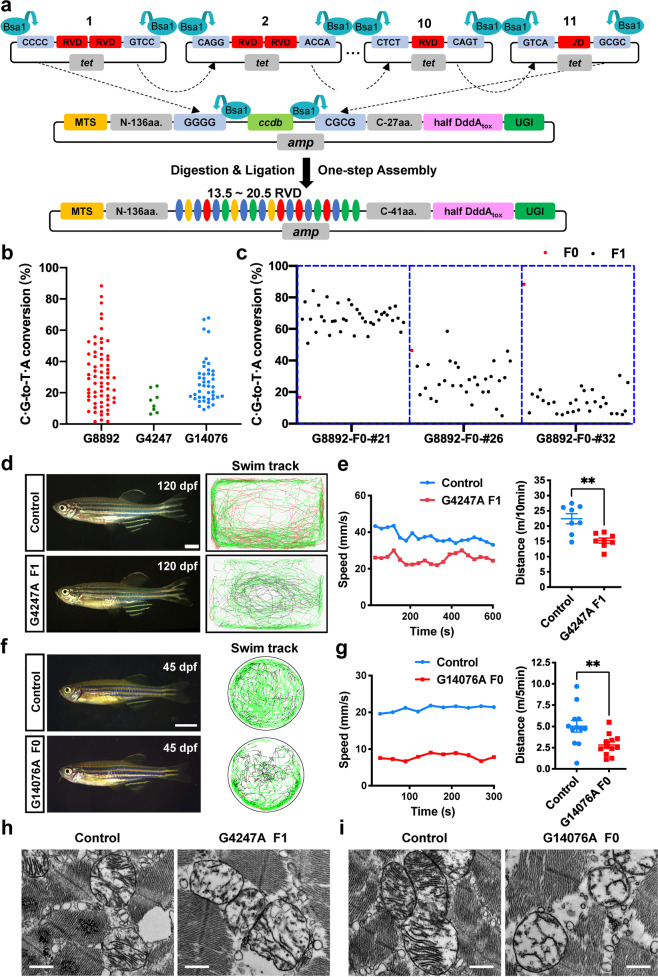
Fig. 1. DdCBE-mediated mtDNA base editing in zebrafish.
a Schematic overview of one-step assembly of DdCBE. Top, plasmids containing intended RVD are cleaved using BsaI to produce sticky ends. Middle, the RVDs with sticky ends are allowed to ligate to backbones sequentially. Bottom, the assembled DdCBE vectors contain 13.5–20.5 RVDs. Twenty-seven amino acids (aa) are put after ccdb in the backbone, and 14 aa are put in half RVD module. There are a total of 41 aa at C-terminus of TALE after assembly. b Deep sequencing analysis of C ∙ G-to-T ∙ A conversions in G8892, G4247 and G14076 founders. c Deep sequencing analysis of transmission of mtDNA mutation in offspring of G8892 founders. Red dot and black dot indicate individual zebrafish of F0 and F1, respectively. d Left panels, live images of control and G4247A F1 zebrafish at 120 days post-fertilization. Lateral view, anterior to the left. Scale bar, 2 mm. Right panels, representative swim tracking of individual animals. Color coding represents movement speed: red, faster than 75 mm/s; green, 25–75 mm/s; black, slower than 25 mm/s. e Quantification of total distance and speed of G4247A F1 zebrafish at 120 days post-fertilization (means ± SEM, n = 8 for each group). Significance was calculated with unpaired two-tailed Student’s t-test (**P < 0.01). f Left panels, live images of control and G14076A founder zebrafish at 45 days post-fertilization. Lateral view, anterior to the left. Scale bar, 2 mm. Right panels, representative swim tracking of individual animals. g Quantification of total distance and speed of G14076A founder zebrafish at 45 days post-fertilization (means ± SEM, n = 12 for each group). Significance was calculated with unpaired two-tailed Student’s t-test (**P < 0.01). h, i TEM images of skeletal muscle from F1 G4247A and F0 G14076A fish demonstrating abnormal mitochondrial ultrastructure. Scale bar, 500 nm.