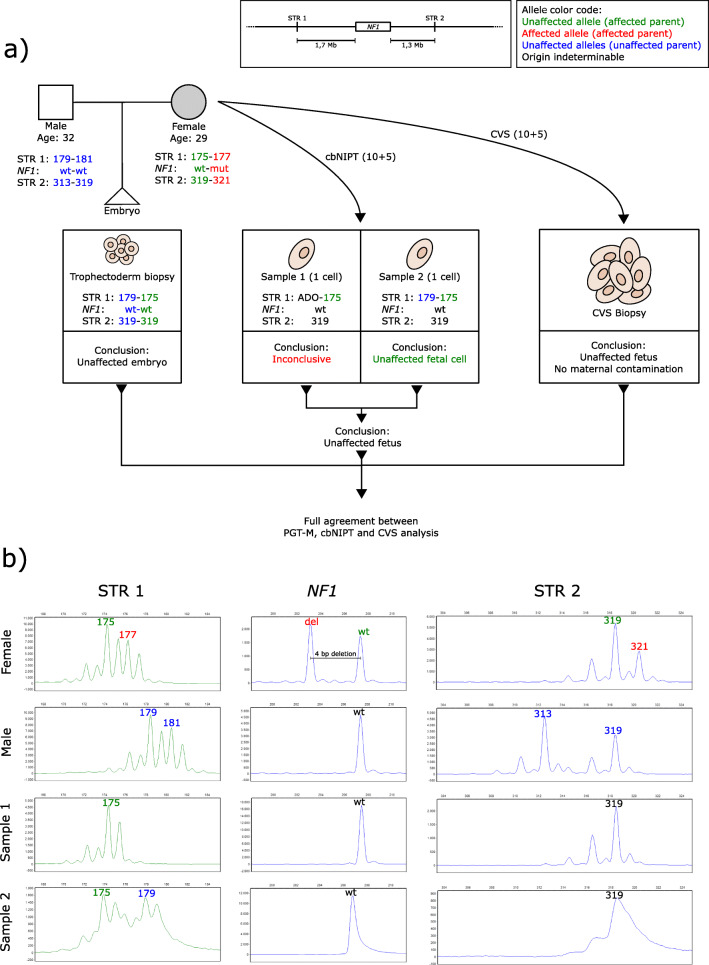
Fig. 1.
Results from case 1. a Flowchart describing the setup and STR markers used for PGT-M and cbNIPT as well as the results and conclusions from PGT, cbNIPT, and CVS. b STR profiles from cbNIPT including paternal and maternal profiles. Insert in the upper right corner details the affected gene and the locations of the STR markers used. Affected alleles are written in red, unaffected in green (the affected parent) or blue (the unaffected parent). Alleles of indeterminable origin are written in black. A couple (29 and 32 years old) seeking PGT due to the female partner being affected by neurofibromatosis type I caused by a deletion (c.7907+4_7del) in the NF1 gene (a). Two STR markers, one fully informative located 1.7 Mb upstream of the NF1 gene (STR 1, NF1-4, see supplementary materials and methods for genomic locations and sequence of self-annotated STR markers) and one semi-informative located 1.3 Mb downstream of the NF1 gene (STR 2, D17S1880), were identified (a insert). Direct mutation detection (by fragment analysis because the mutation is a deletion) coupled with STR analysis was performed on DNA from lysed biopsied trophectoderm cells. An unaffected blastocyst was transferred resulting in pregnancy (a). CVS and blood sampling were performed in gestational week 10+5. Two potential fetal cell samples were isolated from the maternal blood sample (C1-S1 and C1-S2). C1-S2 was classified as inconclusive due to the absence of the paternal allele. C1-S2 was classified as an unaffected fetal cell with the same profile as the transferred embryo. Combined, cbNIPT confirmed the transfer of an unaffected embryo, which was also confirmed by CVS analysis (a). bp, base pair; cbNIPT, cell-based non-invasive prenatal testing; CVS, chorionic villous sampling; Mb, mega bases; PGT, preimplantation genetic testing; PGT-M, PGT for monogenic disorders; STR, short tandem, Cx-Sy, case x sample y