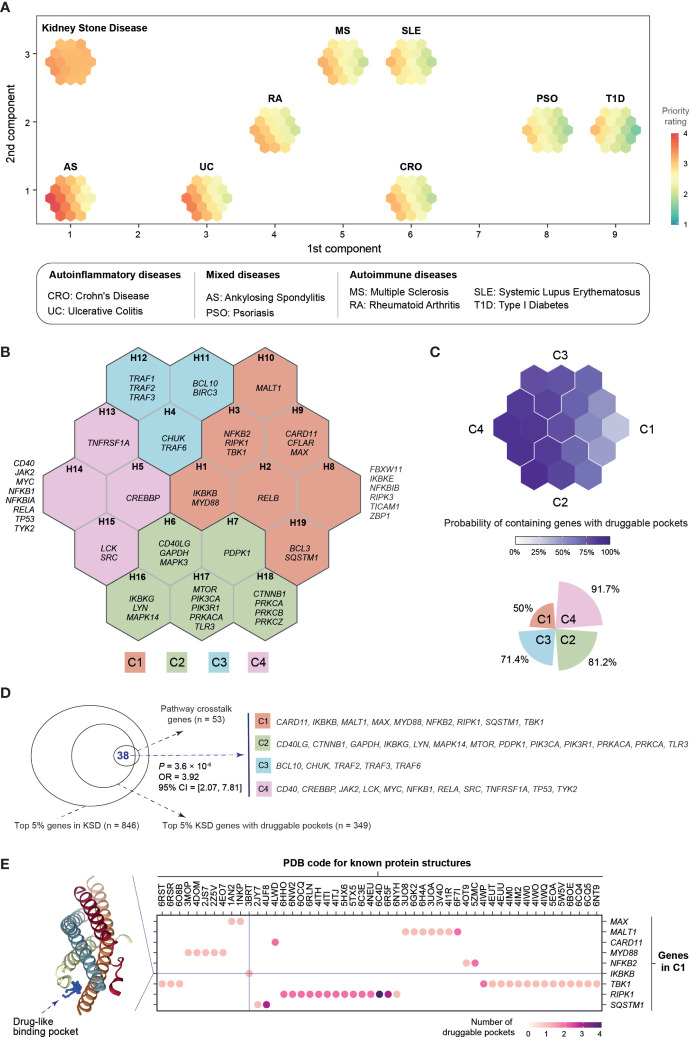
Figure 5.
Cross-disease analysis of pathway crosstalk genes. The target prioritizations for eight immune-related disease traits are sourced from the Pi database. On the basis of degree of autoinflammation versus autoimmunity, these immune traits are grouped into polygenic autoinflammatory diseases (CRO and UC), polygenic autoimmune diseases (MS, RA, SLE and T1D), and mixed diseases having both (AS and PSO). (A) Comparisons using the supra-hexagonal map. This map was learned from the prioritization information of 53 crosstalk genes in kidney stone disease (KSD) and eight immune-related traits. Each map illustrates a trait-specific crosstalk gene prioritization profile. Across traits, genes with similar prioritization patterns are mapped onto the same or nearly position in the map. The outermost frame represents the landscape for the traits analyzed, from which geometric locations of traits delineate their relationships (by the similarity of prioritization profiles between traits). (B) Gene clusters. The map was divided into four target gene clusters (C1–C4) as color coded; each covering continuous hexagons. Hexagons are indexed and expanded outward (H1–H19). Also displayed are genes found in each hexagon. (C) Tractability. The map is color-coded by the probability of each hexagon containing tractable genes, with the percentage of tractable genes summarized for each cluster shown beneath (polar bar). A gene is defined as being tractable if predicted to contain a druggable pocket. (D) Venn diagram illustrating the enrichment for tractable genes. The significance level (P), odds ratio (OR), and 95% confidence intervals (CI) calculated using one-sided Fisher’s exact test. KSD, kidney stone disease. (E) Druggable pockets. Dot plot shows nine tractable genes in C1 (y-axis) and their PDB known protein structures (x-axis). Color-coded is the number of druggable pockets predicted in the structure. An example structure “3BRT” is visualized on the left, with a druggable pocket illustrated using 3D balls in blue.