Abstract
Background
We aimed to construct an artificial intelligence (AI) guided identification of suspicious bone metastatic lesions from the whole-body bone scintigraphy (WBS) images by convolutional neural networks (CNNs).
Methods
We retrospectively collected the 99mTc-MDP WBS images with confirmed bone lesions from 3352 patients with malignancy. 14,972 bone lesions were delineated manually by physicians and annotated as benign and malignant. The lesion-based differentiating performance of the proposed network was evaluated by fivefold cross validation, and compared with the other three popular CNN architectures for medical imaging. The average sensitivity, specificity, accuracy and the area under receiver operating characteristic curve (AUC) were calculated. To delve the outcomes of this study, we conducted subgroup analyses, including lesion burden number and tumor type for the classifying ability of the CNN.
Results
In the fivefold cross validation, our proposed network reached the best average accuracy (81.23%) in identifying suspicious bone lesions compared with InceptionV3 (80.61%), VGG16 (81.13%) and DenseNet169 (76.71%). Additionally, the CNN model's lesion-based average sensitivity and specificity were 81.30% and 81.14%, respectively. Based on the lesion burden numbers of each image, the area under the receiver operating characteristic curve (AUC) was 0.847 in the few group (lesion number n ≤ 3), 0.838 in the medium group (n = 4–6), and 0.862 in the extensive group (n > 6). For the three major primary tumor types, the CNN-based lesion identifying AUC value was 0.870 for lung cancer, 0.900 for prostate cancer, and 0.899 for breast cancer.
Conclusion
The CNN model suggests potential in identifying suspicious benign and malignant bone lesions from whole-body bone scintigraphic images.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12880-021-00662-9.
Keywords: Bone scintigraphy, Bone metastasis, Artificial intelligence, Convolutional neural network
Background
Bone metastasis commonly appears in the advanced stages of cancers [1–4]. It seriously affects the survival quality of patients due to the occurrence of adverse skeletal-related events [2, 5, 6]. The early diagnosis of bone metastasis is beneficial to make appropriate and timely treatment of metastatic bone disease, which can improve the quality of survival [7–10]. Even after the advent of single-photon emission computed tomography combined with computed tomography (SPECT/CT), whole-body bone scintigraphy (WBS) is a standard method to survey the existence and extent of bone metastasis [11]. However, the image resolution and the specificity of WBS are lacking [12]. And the interpretation of WBS is an experience-dependent work and the diagnostic agreement of inter-observer is not satisfactory [13].
Previously, we had proposed an automated diagnostic system of bone metastasis based on multi-view bone scans using an attention-augmented deep neural network [14, 15]. While it achieved considerable accuracy in the patient-based diagnosis from WBS images, a definitive diagnosis for suspicious bone metastatic lesions is still crucial for pragmatic decisions, such as precise bone biopsy, bone surgery and external beam radiotherapy [16]. Thus, a new artificial intelligence (AI) model with lesion-based diagnosis from the WBS image is more valuable for the clinic. Therefore, we fed a fully annotated WBS images dataset to construct a new AI model and evaluated its lesion-based performance in automatic diagnosing suspicious bone metastatic lesions.
Methods
This retrospective single-center study was approved by the Institutional Ethics Committee of West China Hospital of Sichuan University. The written informed consent was waived from the Institutional Ethics Committee of West China Hospital of Sichuan University.
Data resource
The WBS images of patients who were identified lung cancer, prostate cancer and breast cancer were retrieved from our hospital database within the period from Feb. 2012 to Apr. 2019. The WBS was performed using two gamma cameras (GE Discovery NM/CT 670 and Philips Precedence 16 SPECT/CT). The patient received 555 to 740 MBq of technetium-99 m methylene diphosphonate (99mTc-MDP; purchased from Syncor Pharmaceutical Co., Ltd, Chengdu, China) by intravenous injection, and the anterior and posterior views WBS images were obtained approximately 3 h post-injection. The gamma cameras were equipped with low-energy, high-resolution, parallel-hole collimators. The scan speed was 16–20 cm/min, and the matrix size was 512 × 1024. Energy peak was centered at 140 keV with 15% to 20% windows.
The visible bone lesion in WBS images was manually delineated by human experts and annotated into malignant and benign according to the following criteria [17, 18]:
Malignant: bone lesion with increased 99mTc-MDP were identified as malignant (1) when computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography-computed tomography (PET/CT), etc. presented bone destruction; (2) when it appeared newly but couldn’t be ruled out as malignant in follow-up bone scan; (3) when it presented flare phenomenon; (4) when it enlarged and thickened significantly after at least 3 months follow-up.
Benign: bone lesion with increased 99mTc-MDP were identified as benign (1) when CT, MRI and PET/CT, etc. demonstrated fracture, bone cyst, osteogeny, osteophyte, bone bridge, degenerative osteoarthrosis; (2) when it appeared around the bone joint; (3) when it confirmed as trauma.
The diagram of manual delineation and annotation was shown in Fig. 1. Additionally, the patient-based WBS image was assigned to malignant once a lesion was identified as malignant. Finally, from the 3352 patients, 14,972 visible bone lesions were identified as benign or malignant. According to the total number of lesions per WBS image [19], we divided all cases into three groups: few lesions group: 1–3 lesions; medium lesions group: 4–6 lesions; extensive lesions group: > 6 lesions.
Fig. 1.
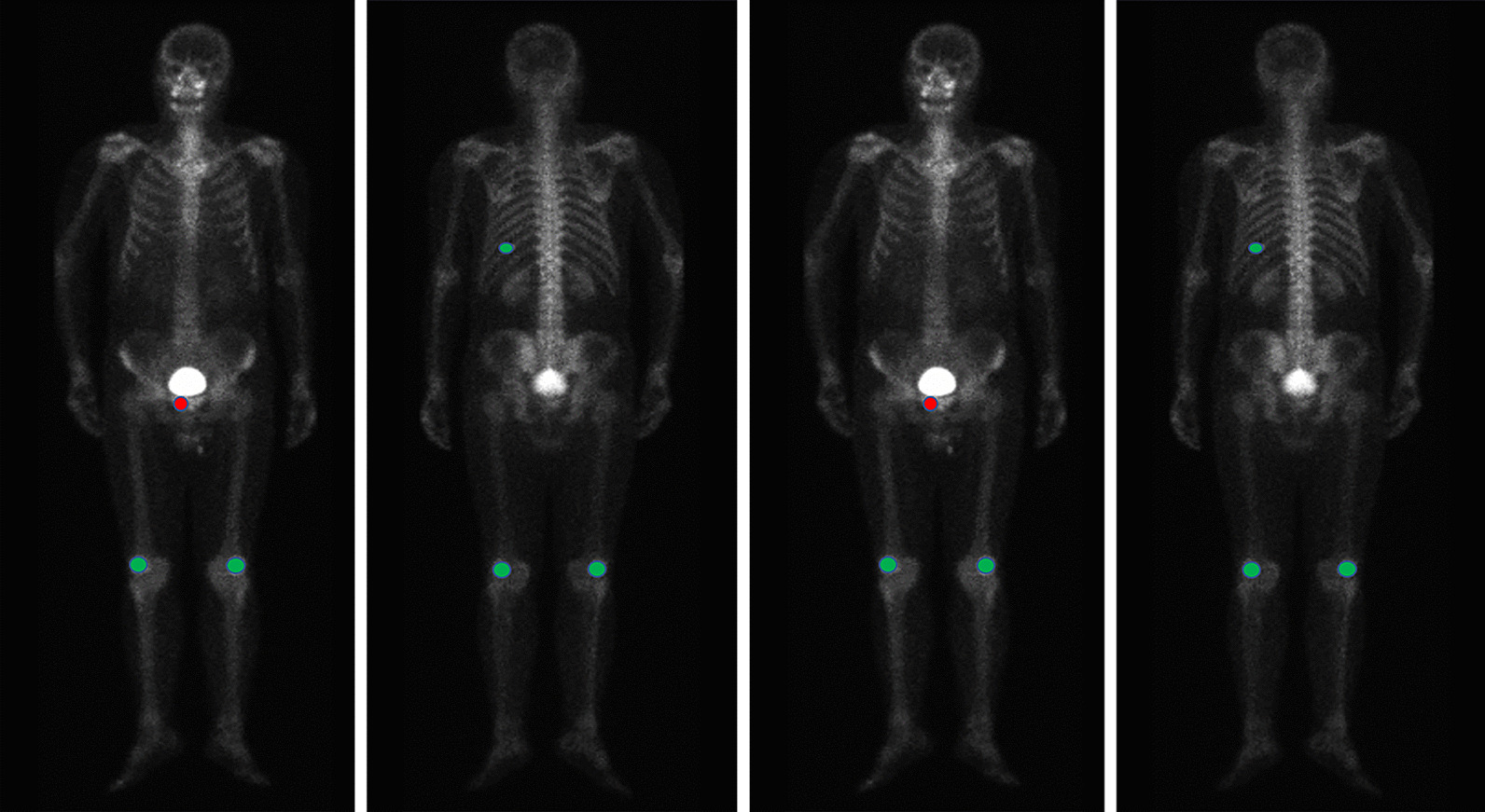
The diagram of manual annotation in WBS image. All visible bone lesions were delineated and annotated as benign and malignant. Red areas represent malignant lesions, while green areas represent benign lesions
Model architecture
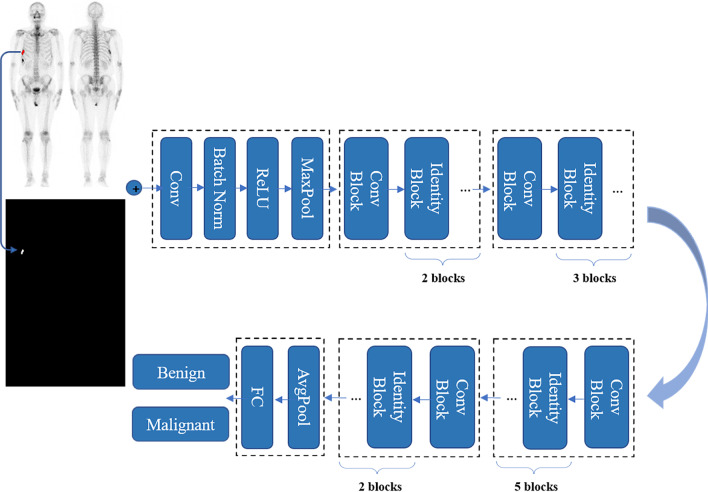
We implemented 2D CNN to automatically identification of bone metastatic lesions. Our network is based on the architecture of ResNet50 [20]. The CNN model was pre-trained on ImageNet, and fine-tuned on our own dataset. Before training the network, a pre-processing step was performed for data curation. The WBS and corresponding lesion mask were resized to 512 × 256. Considering the diagnosis of bone lesions was tremendously correlated to the location and burden extent, we stacked the full-sized images and the corresponding lesion mask on channel, instead of only inputting ROI of lesions. The data consisted of the original WBS image, the corresponding lesion mask and the qualitative of the lesion was used for CNN training. The fivefold cross validation was performed for evaluating the ability of the trained network model to achieve the qualitative task of bone scan lesions. Additionally, three state-of-the-art CNNs that included Inception V3 [21], VGG16 [22] and DenseNet169 [23] were compared with the proposed network.
The developed network was implemented using PyTorch [24], and trained using Adam [25] as the optimizer with a learning rate of 0.001 for 300 epochs. The mini-batch size was fixed 8. During the training process, random horizontal flipping with a probability of 0.5 was applied to the input to increase the diversity of the data. The detailed network architecture is shown in Fig. 2.
Fig. 2.
Architecture of convolutional neural network for AI model
Statistical analysis
The performance of AI was evaluated using diagnostic sensitivity, specificity, accuracy, positive predictive value (PPV), negative predictive value (NPV) and the area under the receiver operating characteristic curve (AUC). The Chi-square test was performed to compare differences in the AI performance between different number of lesions and different primary tumor types. The confusion matrix showed the numbers of true positive, true negative, false positive and false negative. All analyses were conducted using statistical software SPSS22.0 (SPSS Inc, Chicago, Illinois, USA). P values less than 0.05 were considered statistically significant.
Results
Baseline characteristics of patients
3352 cancer patients (Age: 61.61 ± 12.69y; Gender: 1758 males and 1594 females) were retrospectively included in the study and 43.85% of all patients presented bone metastasis. A total of 14,972 visible bone lesions were recognized in all WBS images and 51.23% of them were identified metastasis. The lesion-based metastasis rate was 50.13% in lung cancer, 57.39% in prostate cancer, and 44.61% in breast cancer, respectively. The detailed information was listed in Table 1.
Table 1.
The summary of patient-based and lesion-based analysis in all WBS images
| Lung cancer | Prostate cancer | Breast cancer | Total | |
|---|---|---|---|---|
| Patient-based | ||||
| Total | 1253 | 1017 | 1082 | 3352 |
| Malignant | 567 | 466 | 437 | 1470 |
| Benign | 686 | 551 | 645 | 1882 |
| Metastasis rate | 45.25% | 45.82% | 40.39% | 43.85% |
| Lesion-based | ||||
| Total | 4937 | 5623 | 4412 | 14,972 |
| Malignant | 2475 | 3227 | 1968 | 7670 |
| Benign | 2462 | 2396 | 2444 | 6812 |
| Metastasis rate | 50.13% | 57.39% | 44.61% | 51.23% |
The performance of the proposed network
After fivefold cross validation, the CNN model demonstrated an average sensitivity, specificity, accuracy, PPV and NPV for all visible bone lesions were 81.30%, 81.14%, 81.23%, 81.89% and 80.61%, respectively. When compared with the other three start-of-art CNNs, our proposed network achieved the best accuracy in identification the bone lesions at bone scintigraphy (Tables 2, 3).
Table 2.
The fivefold cross validation results of the proposed network
| Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | |
|---|---|---|---|---|---|
| Number of patients (benign/malignant) | 669 (376/293) | 669 (376/293) | 671 (376/295) | 669 (376/293) | 674 (378/296) |
| Number of lesions (benign/malignant) | 2986 (1505/1481) | 2952 (1389/1563) | 3084 (1468/1616) | 2897 (1475/1422) | 3053 (1465/1588) |
| Sensitivity | 83.19 | 83.11 | 79.89 | 76.79 | 83.5 |
| Specificity | 78.07 | 82.58 | 82.7 | 82.78 | 79.59 |
| Accuracy | 80.61 | 82.86 | 81.23 | 79.84 | 81.62 |
| PPV | 78.87 | 84.3 | 83.56 | 81.13 | 81.6 |
| NPV | 82.51 | 81.29 | 78.88 | 78.72 | 81.65 |
Table 3.
The comparison of the proposed network and other three networks
| Sensitivity | Specificity | Accuracy | PPV | NPV | |
|---|---|---|---|---|---|
| Our model | 81.30 | 81.14 | 81.23 | 81.89 | 80.61 |
| InceptionV3 | 77.29 | 84.02 | 80.61 | 83.53 | 78.00 |
| VGG16 | 78.73 | 83.51 | 81.13 | 83.39 | 79.14 |
| DenseNet169 | 67.90 | 85.73 | 76.71 | 83.17 | 72.16 |
Subgroup analysis of proposed network
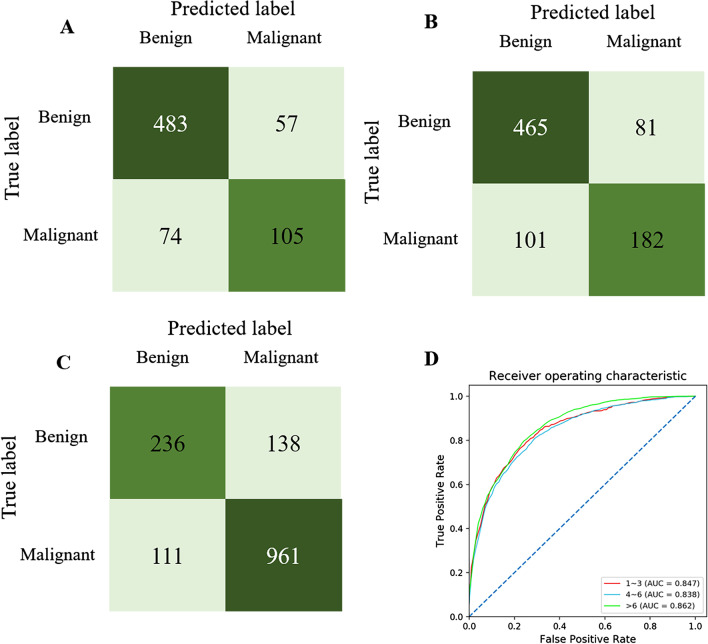
Based on the number of lesions per image, we found that the AI model reached the highest sensitivity (89.56%, P < 0.001), accuracy (82.79%, P = 0.018) and PPV (87.37%, P < 0.001) in the extensive lesions group as shown in Table 4. Whereas, the highest specificity (89.41%, P < 0.001) and NPV (86.76%, P < 0.001) of the AI model were captured in few lesions group. We also calculated the AUC to evaluate the diagnostic performance of the AI model, which was 0.847 in the few lesions group, 0.838 in the medium lesions group, and 0.862 in the extensive lesions group. And the confusion matrix directly demonstrated the true labels and predicted labels in the three groups (Fig. 3).
Table 4.
The lesion-based diagnostic performance of AI model in testing cohort and the comparison of the AI performance among few, medium and extensive lesion groups
| Group for number of lesions | χ2 | P value | |||
|---|---|---|---|---|---|
| Few | Medium | Extensive | |||
| Sensitivity | 58.63 | 64.34 | 89.56 | 163.41 | < 0.001 |
| Specificity | 89.41 | 85.24 | 62.85 | 108.69 | < 0.001 |
| Accuracy | 81.78 | 78.03 | 82.79 | 8.06 | 0.018 |
| PPV | 64.89 | 69.67 | 87.37 | 83.70 | < 0.001 |
| NPV | 86.76 | 82.04 | 67.93 | 49.24 | < 0.001 |
Chi-square test was performed to compare the performance of AI model among different groups of the number of lesions. Few lesions group: 1–3 lesions per image; Medium lesions group: 4–6 lesions per image; Extensive lesions group: > 6 lesions per image
Fig. 3.
The confusion matrix of few lesions group (A), medium lesions group (B), extensive lesions group (C). The ROC of the three groups in the lesion-based diagnosis (D)
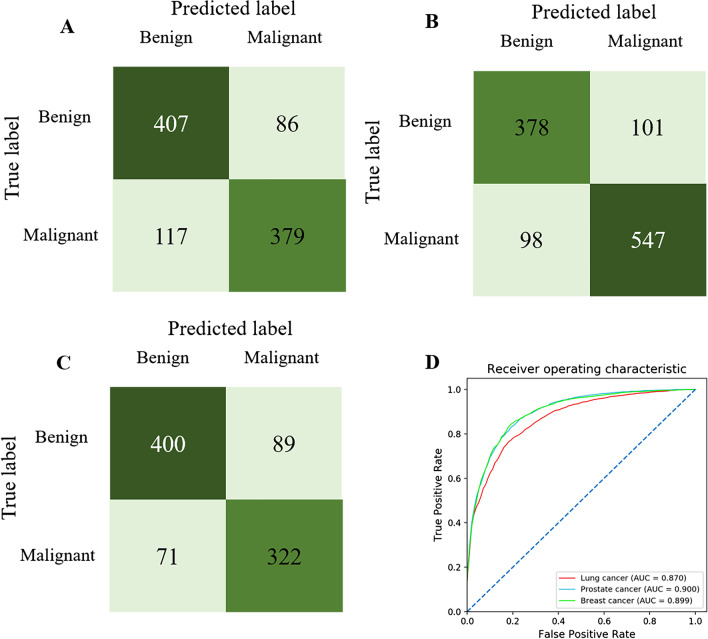
The detailed results based on the primary tumor types were shown in Table 5, the results demonstrated the highest diagnostic sensitivity (84.66%, P = 0.002) in the prostate cancer group. Albeit slightly higher accuracy (82.30%) in the prostate cancer group, there was no statistical significance (P = 0.209) comparing with the lung cancer group (79.40%) and breast cancer group (81.82%). The specificity in lung cancer (82.52%), prostate cancer (79.07%) and breast cancer (81.78%) group also did not indicate statistical significance between each other (P = 0.354). Furthermore, the AUC was 0.870 for lung cancer, 0.900 for prostate cancer, 0.899 for breast cancer. The confusion matrix directly demonstrated the true labels and predicted labels in the three groups (Fig. 4).
Table 5.
The lesion-based diagnostic performance of AI model in the testing cohort and the comparison of the AI performance among lung, prostate and breast cancers
| Group for primary tumor types | χ2 | P value | |||
|---|---|---|---|---|---|
| Lung cancer | Prostate cancer | Breast cancer | |||
| Sensitivity | 76.16 | 84.66 | 81.65 | 12.88 | 0.002 |
| Specificity | 82.52 | 79.07 | 81.78 | 2.08 | 0.354 |
| Accuracy | 79.40 | 82.30 | 81.82 | 3.13 | 0.209 |
| PPV | 81.12 | 84.43 | 78.33 | 6.52 | 0.038 |
| NPV | 77.70 | 79.52 | 85.05 | 8.85 | 0.012 |
Chi-square test was performed to compare the performance of AI model among different tumor types
Fig. 4.
The confusion matrix of lung cancer group (A), prostate cancer group (B), breast cancer group (C). The ROC of the three groups in the lesion-based diagnosis (D)
Additionally, we also evaluated the lesion-based diagnostic performance of the AI model according to the different number of lesions per image (few, medium and extensive lesions group) in lung cancer, prostate cancer and breast cancer, respectively. The results were supported as Additional file 1: Table 1 and Additional file 2: Figs. 1, 2, and 3.
Discussion
The definitive identification of abnormal bone lesions is beneficial to proper personalized treatment and subserves the patients who were suffering from advanced malignant cancers [26]. However, the precise differentiation of suspicious bone lesions is still tricky based on the WBS images only [27]. In light of the superiority of artificial intelligence in image feature extraction and big data analysis, we developed a new AI model using the 2D CNN to explore the potential for automatically definitive identification of suspicious bone metastatic lesions from WBS images.
In general, our AI model achieved moderate performance in the identification of suspicious bone lesions with a sensitivity of 81.30% and specificity of 81.14%. We found that AI indicated significantly higher accuracy in the extensive-lesions group (n > 6, accuracy = 82.79%) than that in the few-lesions group (n ≤ 3, accuracy = 81.78%, p < 0.05) and medium-lesions group (n = 4–6, accuracy = 78.03%, p < 0.05), this might be beneficial from the deep neural network which imitating human thinking model. Originally, classification of every single lesion is judged independently, regardless of the other lesions that appeared in the same image. However, nuclear medicine physicians usually take other lesions and additional cues into account when determining one single lesion itself. For example, an isolated lesion without other nearby lesions would be more difficult to assert benign or malignant, while multiple lesions that occur within a narrow region would be more likely malignant. We input corresponding lesion masks to the CNN and take the whole WBS image into account, and this might be a possible reason for the improved accuracy of the extensive-lesions group.
Previous studies also reported AI for bone lesion identification from WBS images. The authors used a ladder network to pre-train a nerual network with an unlabeled dataset [28]. On the metastasis classification task, It reached a sensitivity of 0.657 and a specificity of 0.857. Another similar study also build a model to detect and identify bone metastasis from bone scintigraphy images through negative mining, pre-training, the convolutional neural network, and deep learning [29]. The mean lesion-based sensitivity and precision rates for bone metastasis classification were 0.72 and 0.90, respectively. In our study, the lesion-based sensitivity, specificity and precision values for metastasis classification were 0.813, 0.811 and 0.819, respectively. It is difficult to compare the difference of algorithms, all studies have used in-house datasets of a gold standard and these datasets were not open. We were not able to try other datasets using our algorithm. Therefore, the performances reported by other researchers can only be used as references, rather than for objective comparison. It is worth mentioning that the aforementioned AI was focused on the chest image instead of the whole body. This strategy excluded the influence from keen osteoarthritis, degenerative changes of lumbar/cervical vertebrae, but it was limited to analyzing the metastases in other regions such as the pelvis, sacrum, iliac joints and other distant lesions. Addittionaly, we stacked the WBS and the corresponding lesion mask in channel and input it into the network. Thus, this CNN approach could select any suspicious bone lesion that needs to be input manually and obviate missed lesion detection and wrong lesion detection.
Three common kinds of primary cancers were investigated in this study. The different sensitivity among primary cancer types seemed to be affiliated to osteoblastic and osteolytic activity. The highest sensitivity appeared in the prostate cancer group and it is consistent with other former studies [17]. The probable reason is due to the typical osteoblastic metastasis principally in prostate cancer, though it is also associated with the osteoclastic process and bone resorption [30]. On the other hand, lung cancer and breast cancer group showed more significant osteolytic changes and corresponding mild radioactivity in lesions [31, 32].
Generally, our AI model achieved a moderate accuracy, sensitivity and specificity in the lesion-based diagnosis of WBS images, the false-positive lesions and false-negative lesions still could not be avoided. It is limited to the substantive character and specificity of 99mTc-MDP imaging technology. Most pathological bone conditions, whether of infectious, traumatic, neoplastic or other origin could demonstrate as an increased radioactive signal in WBS images [33]. There are still several limitations in the current study. Firstly, since it is impossible to obtain the pathological result of each lesion, we made the “gold labels” based on the patients’ medical records, the follow-up bone scans, CT, MRI, PET/CT images, etc., which may not be totally correct for every lesion. Secondly, the labeled lesions on WBS images were all visible, which means only the “hotspots” were included, whereas some “cold lesions” were missed. Then, at present, this AI model was constructed by those non-quantitative images, the indraught of anatomical localization parameter and quantitative index might further improve the property, all of which would be paid attention in our future studies. Even though the AI model is not always correct, it still can be used by nuclear medicine physicians for assisting the bone lesions analysis and the final interpretation of an examination, especially for the patients who could not be performed SPECT/CT timely due to the poverty of resource devices.
Conclusions
The AI model based on CNN reached a moderate lesion-based performance in the diagnosis of suspicious bone metastatic lesions from WBS images. Even though the AI model is not always correct, it could serve as an effective auxiliary tool for diagnosis and guidance in patients with suspicious bone metastatic lesions in daily clinical practice.
Supplementary Information
Additional file 1. The lesion-based diagnostic performance of the AI model according to the different number of lesions per image (few, medium and extensive lesions group) in lung cancer, prostate cancer and breast cancer.
Additional file 2: Fig. S1. The confusion matrix of few lesions group (A), medium lesions group (B), extensive lesions group (C) in lung cancer group. The ROC of the three groups in the lesion-based diagnosis (D).
Additional file 3: Fig. S2. The confusion matrix of few lesions group (A), medium lesions group (B), extensive lesions group (C) in prostate cancer group. The ROC of the three groups in the lesion-based diagnosis (D).
Additional file 4: Fig. S3. The confusion matrix of few lesions group (A), medium lesions group (B), extensive lesions group (C) in breast cancer group. The ROC of the three groups in the lesion-based diagnosis (D).
Acknowledgements
This project was financially supported by the National Major Science and Technology Projects of China (2018AAA0100201), the Sichuan Science and Technology Program of China (2020JDRC0042) and “1.3.5 project for disciplines of excellence in West China Hospital (ZYGD18016 & 2021HXFH033).
Abbreviations
- AI
Artificial intelligence
- WBS
Whole-body bone scintigraphy
- AUC
Area under receiver operating characteristic curve
- SPECT/CT
Single-photon emission computed tomography combined with computed tomography
- CT
Computed tomography
- MRI
Magnetic resonance imaging
- PET/CT
Positron emission tomography-computed tomography
- CNN
Convolution neural networks
- PPV
Positive predictive value
- NPV
Negative predictive value
Authors' contributions
Conceptualization, CH, ZZ, YP and LY; methodology, PY and WJ; software, PY, WJ and ZY; validation, WJ, CJ and ZX; formal analysis, LY and YP; investigation, YP and XY; resources, LL and ZY; data curation, XY, JL, and YP; writing—original draft preparation, LY, YP and PY; writing—review and editing, CH and ZZ; visualization, CH; supervision, LL, CH and ZY; project administration, ZZ, LL; funding acquisition, CH, PY, ZY, and LL. All authors have read and agreed to the published version of the manuscript.
Funding
This project was financially supported by the National Major Science and Technology Projects of China (2018AAA0100201), the Sichuan Science and Technology Program of China (2020JDRC0042) and “1.3.5 project for disciplines of excellence in West China Hospital (ZYGD18016 & 2021HXFH033).
Availability of data and materials
The datasets generated and analyzed during the current study are not publicly available but available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study has been performed in accordance with the Declaration of Helsinki and has been approved by the Institutional Ethics Committee of West China Hospital in Sichuan University. As this study was of retrospective nature, a consent form was waived by the Institutional Ethics Committee of West China Hospital in Sichuan University.
Consent for publication
Not applicable.
Competing interests
The authors declare there they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yemei Liu and Pei Yang contributed equally to this project
Contributor Information
Huawei Cai, Email: hw.cai@yahoo.com.
Zhen Zhao, Email: zhaozhen1982@126.com.
References
- 1.Tsuya A, Kurata T, Tamura K, Fukuoka M. Skeletal metastases in non-small cell lung cancer: a retrospective study. Lung Cancer. 2007;57:229–232. doi: 10.1016/j.lungcan.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Coleman RE. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin Cancer Res. 2006;12:6243s–6249s. doi: 10.1158/1078-0432.CCR-06-0931. [DOI] [PubMed] [Google Scholar]
- 3.Tantivejkul K, Kalikin LM, Pienta KJ. Dynamic process of prostate cancer metastasis to bone. J Cell Biochem. 2004;91:706–717. doi: 10.1002/jcb.10664. [DOI] [PubMed] [Google Scholar]
- 4.Schneider C, Fehr MK, Steiner RA, Hagen D, Haller U, Fink D. Frequency and distribution pattern of distant metastases in breast cancer patients at the time of primary presentation. Arch Gynecol Obstet. 2003;269:9–12. doi: 10.1007/s00404-002-0445-x. [DOI] [PubMed] [Google Scholar]
- 5.Sun JM, Ahn JS, Lee S, Kim JA, Lee J, Park YH, Park HC, Ahn MJ, Ahn YC, Park K. Predictors of skeletal-related events in non-small cell lung cancer patients with bone metastases. Lung Cancer. 2011;71:89–93. doi: 10.1016/j.lungcan.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Al Husaini H, Wheatley-Price P, Clemons M, Shepherd FA. Prevention and management of bone metastases in lung cancer: a review. J Thorac Oncol. 2009;4:251–259. doi: 10.1097/JTO.0b013e31819518fc. [DOI] [PubMed] [Google Scholar]
- 7.Owari T, Miyake M, Nakai Y, Hori S, Tomizawa M, Ichikawa K, Shimizu T, Iida K, Samma S, Iemura Y, et al. Clinical benefit of early treatment with bone-modifying agents for preventing skeletal-related events in patients with genitourinary cancer with bone metastasis: a multi-institutional retrospective study. Int J Urol. 2019;26:630–637. doi: 10.1111/iju.13939. [DOI] [PubMed] [Google Scholar]
- 8.Hirai T, Shinoda Y, Tateishi R, Asaoka Y, Uchino K, Wake T, Kobayashi H, Ikegami M, Sawada R, Haga N, et al. Early detection of bone metastases of hepatocellular carcinoma reduces bone fracture and paralysis. Jpn J Clin Oncol. 2019;49:529–536. doi: 10.1093/jjco/hyz028. [DOI] [PubMed] [Google Scholar]
- 9.Heeke A, Nunes MR, Lynce F. Bone-modifying agents in early-stage and advanced breast cancer. Curr Breast Cancer Rep. 2018;10:241–250. doi: 10.1007/s12609-018-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosen DB, Benjamin CD, Yang JC, Doyle C, Zhang Z, Barker CA, Vaynrub M, Yang TJ, Gillespie EF. Early palliative radiation versus observation for high-risk asymptomatic or minimally symptomatic bone metastases: study protocol for a randomized controlled trial. BMC Cancer. 2020;20:1115. doi: 10.1186/s12885-020-07591-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Love C, Din AS, Tomas MB, Kalapparambath TP, Palestro CJ. Radionuclide bone imaging: an illustrative review. Radiographics. 2003;23:341–358. doi: 10.1148/rg.232025103. [DOI] [PubMed] [Google Scholar]
- 12.Lin Q, Luo M, Gao R, Li T, Man Z, Cao Y, Wang H. Deep learning based automatic segmentation of metastasis hotspots in thorax bone SPECT images. PLoS ONE. 2020;15:e0243253. doi: 10.1371/journal.pone.0243253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadik M, Suurkula M, Höglund P, Järund A, Edenbrandt L. Quality of planar whole-body bone scan interpretations—a nationwide survey. Eur J Nucl Med Mol Imaging. 2008;35:1464–1472. doi: 10.1007/s00259-008-0721-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z, Pi Y, Jiang L, Xiang Y, Wei J, Yang P, Zhang W, Zhong X, Zhou K, Li Y, et al. Deep neural network based artificial intelligence assisted diagnosis of bone scintigraphy for cancer bone metastasis. Sci Rep. 2020;10:17046. doi: 10.1038/s41598-020-74135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pi Y, Zhao Z, Xiang Y, Li Y, Cai H, Yi Z. Automated diagnosis of bone metastasis based on multi-view bone scans using attention-augmented deep neural networks. Med Image Anal. 2020;65:101784. doi: 10.1016/j.media.2020.101784. [DOI] [PubMed] [Google Scholar]
- 16.Harvie P, Whitwell D. Metastatic bone disease: Have we improved after a decade of guidelines? Bone Joint Res. 2013;2:96–101. doi: 10.1302/2046-3758.26.2000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokuda O, Harada Y, Ohishi Y, Matsunaga N, Edenbrandt L. Investigation of computer-aided diagnosis system for bone scans: a retrospective analysis in 406 patients. Ann Nucl Med. 2014;28:329–339. doi: 10.1007/s12149-014-0819-8. [DOI] [PubMed] [Google Scholar]
- 18.Sadik M, Suurkula M, Höglund P, Järund A, Edenbrandt L. Improved classifications of planar whole-body bone scans using a computer-assisted diagnosis system: a multicenter, multiple-reader, multiple-case study. J Nucl Med. 2009;50:368–375. doi: 10.2967/jnumed.108.058883. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Shen Y, Zhu S. Distribution features of skeletal metastases: a comparative study between pulmonary and prostate cancers. PLoS ONE. 2015;10:e0143437. doi: 10.1371/journal.pone.0143437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He K, et al. Identity mappings in deep residual networks. In: European conference on computer vision. Cham: Springer, 2016.
- 21.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: IEEE conference on computer vision and pattern recognition; 2016. p. 2818–26.
- 22.Simonyan K, Zisserman A. VGG-16. arXiv 2014.
- 23.Huang G, Liu Z, Der Maaten LV, Weinberger KQ. Densely connected convolutional networks. In: IEEE conference on computer vision and pattern recognition; 2017. p. 2261–9.
- 24.Paszke A, et al. Automatic differentiation in pytorch. 2017.
- 25.Kingma, Diederik P, Jimmy BA. A method for stochastic optimization. arXiv preprint arXiv:1412.6980.2014.
- 26.Yu S, Jiang Z, Zhang L, Niu X, Wang L, Wu N, Ma J. Chinese expert consensus statement on clinical diagnosis and treatment of malignant tumor bone metastasis and bone related diseases. Chin-Ger J Clin Oncol. 2010;9:1–12. doi: 10.1007/s10330-009-0188-2. [DOI] [Google Scholar]
- 27.Utsunomiya D, Shiraishi S, Imuta M, Tomiguchi S, Kawanaka K, Morishita S, Awai K, Yamashita Y. Added value of SPECT/CT fusion in assessing suspected bone metastasis: comparison with scintigraphy alone and nonfused scintigraphy and CT. Radiology. 2006;238:264–271. doi: 10.1148/radiol.2373041358. [DOI] [PubMed] [Google Scholar]
- 28.Apiparakoon T, Rakratchatakul N, Chantadisai M, Vutrapongwatana U, Kingpetch K, Sirisalipoch S, Rakvongthai Y, Chaiwatanarat T, Chuangsuwanich E. MaligNet: semisupervised learning for bone lesion instance segmentation using bone scintigraphy. IEEE Access. 2020;8:27047–27066. doi: 10.1109/ACCESS.2020.2971391. [DOI] [Google Scholar]
- 29.Cheng DC, Hsieh TC, Yen KY, Kao CH. Lesion-based bone metastasis detection in chest bone scintigraphy images of prostate cancer patients using pre-train, negative mining, and deep learning. Diagnostics (Basel). 2021;11. [DOI] [PMC free article] [PubMed]
- 30.Messiou C, Cook G, deSouza NM. Imaging metastatic bone disease from carcinoma of the prostate. Br J Cancer. 2009;101:1225–1232. doi: 10.1038/sj.bjc.6605334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roato I. Bone metastases: when and how lung cancer interacts with bone. World J Clin Oncol. 2014;5:149–155. doi: 10.5306/wjco.v5.i2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M, Xia F, Wei Y, Wei X. Molecular mechanisms and clinical management of cancer bone metastasis. Bone Res. 2020;8:30. doi: 10.1038/s41413-020-00105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van den Wyngaert T, Strobel K, Kampen WU, Kuwert T, van der Bruggen W, Mohan HK, Gnanasegaran G, Delgado-Bolton R, Weber WA, Beheshti M, et al. The EANM practice guidelines for bone scintigraphy. Eur J Nucl Med Mol Imaging. 2016;43:1723–1738. doi: 10.1007/s00259-016-3415-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The lesion-based diagnostic performance of the AI model according to the different number of lesions per image (few, medium and extensive lesions group) in lung cancer, prostate cancer and breast cancer.
Additional file 2: Fig. S1. The confusion matrix of few lesions group (A), medium lesions group (B), extensive lesions group (C) in lung cancer group. The ROC of the three groups in the lesion-based diagnosis (D).
Additional file 3: Fig. S2. The confusion matrix of few lesions group (A), medium lesions group (B), extensive lesions group (C) in prostate cancer group. The ROC of the three groups in the lesion-based diagnosis (D).
Additional file 4: Fig. S3. The confusion matrix of few lesions group (A), medium lesions group (B), extensive lesions group (C) in breast cancer group. The ROC of the three groups in the lesion-based diagnosis (D).
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available but available from the corresponding author upon reasonable request.