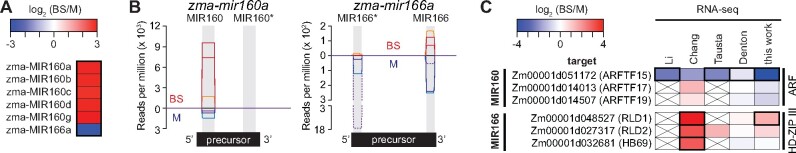
Figure 6.
Differential expression of microRNAs and their TF targets in BS and M fractions. A, Differential expression of microRNAs from sRNA-seq analysis of BS and M fractions. Six genes identified by DESeq2 are shown [|log2 (BS/M)| > 1 and FDR < 0.05]. The mature zma-MIR160 microRNAs have identical sequences, so sRNA-seq cannot distinguish them. B, Normalized read coverage of genes encoding zma-MIR160a and zma-MIR166a. Coverage was normalized by million reads mapped to all microRNA precursors. Single sequence reads spanned the full length of each microRNA, resulting in uniform coverage across their length. Each line represents one replicate, with reads from the BS and M fractions shown above and below the horizontal line, respectively. Each microRNA precursor (diagrammed at bottom) folds into a hairpin via the pairing of complementary regions (gray). One strand in the duplex becomes mature microRNA and the other is typically degraded. An asterisk marks the degraded strand identified by miRBase (http://www.mirbase.org/). C, Cell-type-specific expression of mRNA targets of differentially expressed microRNAs. The mRNA targets of MIR166 and MIR160 were recovered from the DPMIND database (Fei et al., 2018). The original log2 (BS/M) values reported in prior RNA-seq datasets are summarized. Genes reported as differentially expressed are bordered in black. Crossed boxes indicate absence of data. ARFTF, auxin-responsive factor TF; HD-ZIP III, Class III homeodomain leucine zipper; RLD1/2, Rolled leaf1/2; HB69, homeobox-transcription factor69.