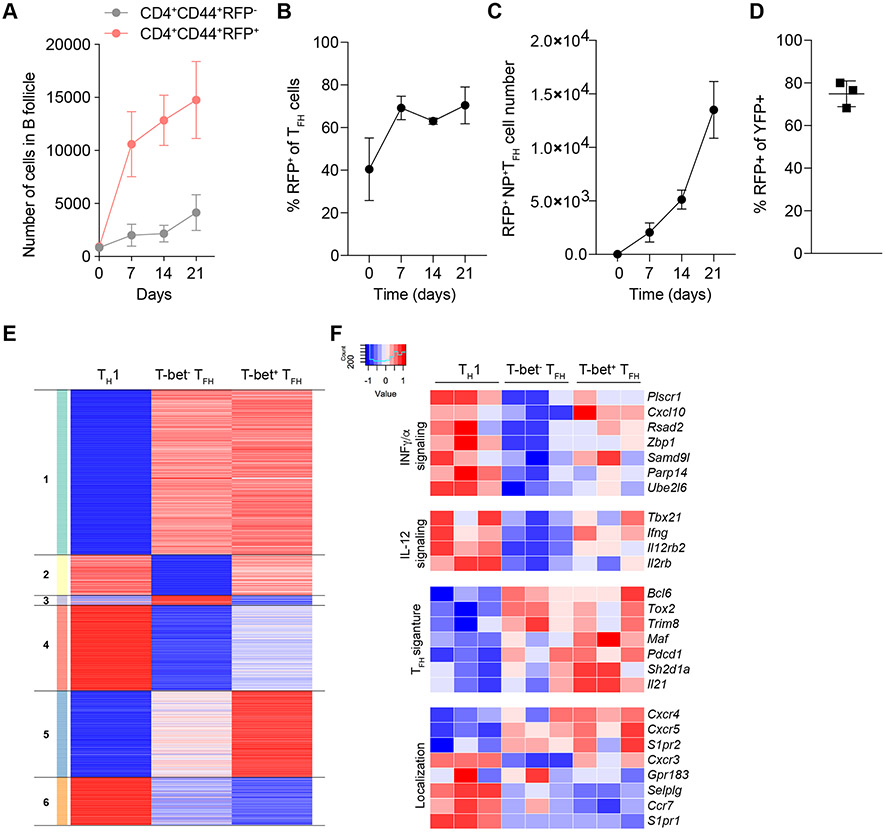
Figure 6. T-bet+ TFH cells are induced during viral infection are a stable population functionally distinct from T-bet− TFH.
(A to C) Tbx21-Cre mice were intranasally infected with 50 TCID50 influenza (PR8). Mediastinal lymph nodes were collected on indicated time points and analyzed by confocal microscopy and flow cytometry. (A) Cell numbers of CD4+CD44+RFP+ and CD4+CD44+RFP− in B cell follicles. (B) Percent RFP+ of CD4+CD44+CXCR5HiPD-1Hi T cells analyzed by confocal microscopy. (C) Numbers of NP tetramer+CD4+CD44+CXCR5HiPD-1HiRFP+ T cells. (D) Tbx21tdTomato-T2A-creERT2Rosa26Lox-STOP-Lox-YFP mice were intranasally infected with 50 TCID50 influenza (PR8); 7 days after infection mice were gavaged with 1 dose of tamoxifen. Mice were analyzed on day 14 after infection. Percent RFP+ of YFP+CD4+CD44+CXCR5HiPD-1Hi T cells. (E and F) Tbx21-Cre; Bcl6-YFP/Bcl6WT; Foxp3Thy1.1 mice were intranasally infected with 50 TCID50 influenza (PR8). Mediastinal lymph nodes were collected on day 14. Indicated cell types were sorted and analyzed by RNA-seq. (E) Heatmap showing k-means clustering of genes differentially expressed in any pairwise comparison. Values are Z-score normalized by row. (F) Expression patterns of select IFN and IL-12 signaling, TFH signature and cell migration-related genes. (A to C) The data are shown as mean ± s.e.m. (D) Each point represents an individual mouse with mean ± s.e.m. All data are representative of ≥ 2 experiments, n ≥ 3 mice per group. (E and F) RNA-seq analysis of gene expression was performed using three biological replicates.