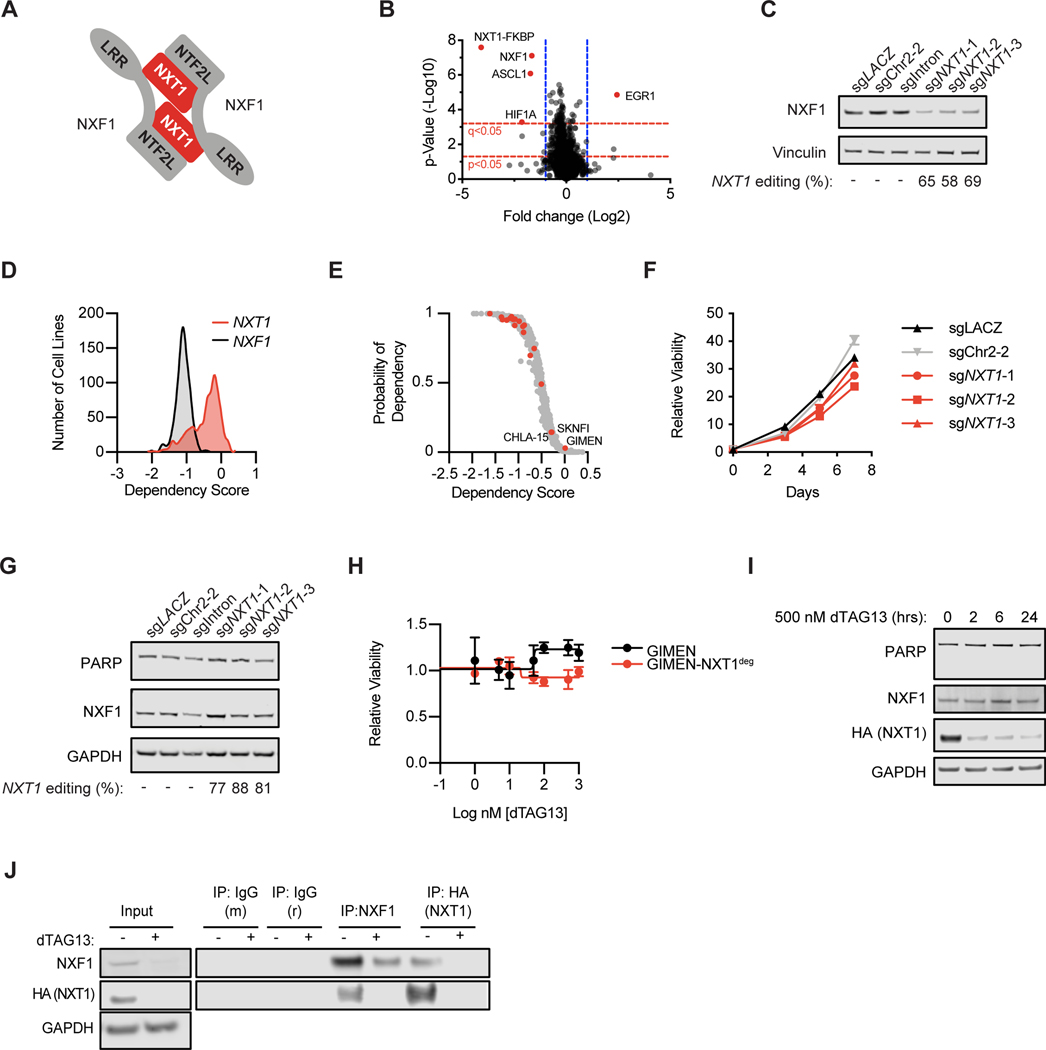
Figure 3. NXT1 loss leads to loss of NXF1.
A, Diagram of NXT1 and NXF1 heterodimer (adapted from Aibara et al. (23)). B, Quantitative proteomics after 2 hours of dTAG13 treatment in KELLY-NXT1deg. X-axis indicates log2 of the relative protein abundance of mean dTAG13-treated to mean control (DMSO) samples. Y-axis indicates the -log10 of the p-value. Red dotted lines indicate q- and p-value cut-offs as indicated, and blue dotted lines indicate a cut-off of log2 fold change of 1. Proteins that reach these cut-offs are labeled. C, Western blot of NXF1 and GAPDH levels in SK-N-BE(2) cells five days after infection with constitutive CRISPR guides as indicated. Genetic editing of the NXT1 locus, calculated using TIDE, is shown below. D, Spline curves of dependency score distribution for all 739 cell lines in the DepMap data set. In red is NXT1 and in black is NXF1. Dependency scores are calculated with CERES and scaled such that −1 is equivalent to pan-essentials, and 0 is equivalent to negative control guides. E, NXT1 dependency in the DepMap dataset for neuroblastoma and other cell lines is shown. On the y-axis is the probability of dependency on NXT1 for each cell line, and on the x-axis is the dependency score calculated by CERES. Each point represents an individual cancer cell line. In red, neuroblastoma cell lines (n=19) are shown. In gray, other cell lines (n=720) are shown. Three neuroblastoma cell lines that do not show dependency on NXT1 are labeled with their cell line names. F, GIMEN cells were infected with negative control (sgLACZ or sgChr2–2, black and gray) and three different sgRNAs targeting NXT1 (red). Viability relative to day 0 was determined using CellTiter-Glo on the indicated days and mean +/− stdev of replicates is shown. G, GIMEN cells were stably infected with indicated sgRNAs and a western blot assessing NXF1, PARP, and GAPDH levels was performed. Below, the percent of CRISPR editing at the NXT1 locus calculated with the TIDE algorithm is shown. H, Dose-response curve for GIMEN parental (black) and GIMEN-NXT1deg (red) where endogenous NXT1 has been knocked out, and a degradable exogenous form of NXT1 is expressed (see methods). I, Western blot showing PARP, NXF1, HA-NXT1, and GAPDH levels after treatment with 500 nM dTAG13 for the indicated amount of time in GIMEN-NXT1deg. J, Immunoprecipitations were performed using antibodies targeting mouse and rabbit IgG, NXF1 (mouse) and HA (rabbit) six hours after DMSO or 500 nM dTAG13 treatment, as indicated in the KELLY-NXT1deg cell line. NXF1 and HA levels are shown detecting endogenous NXF1 and HA-tagged NXT1 respectively. Input lysate is shown to the left with NXF1, HA, and GAPDH.