Figure 5. Silencing of ALDH2 in human AML cell lines and patient samples is associated with promoter DNA hypermethylation.
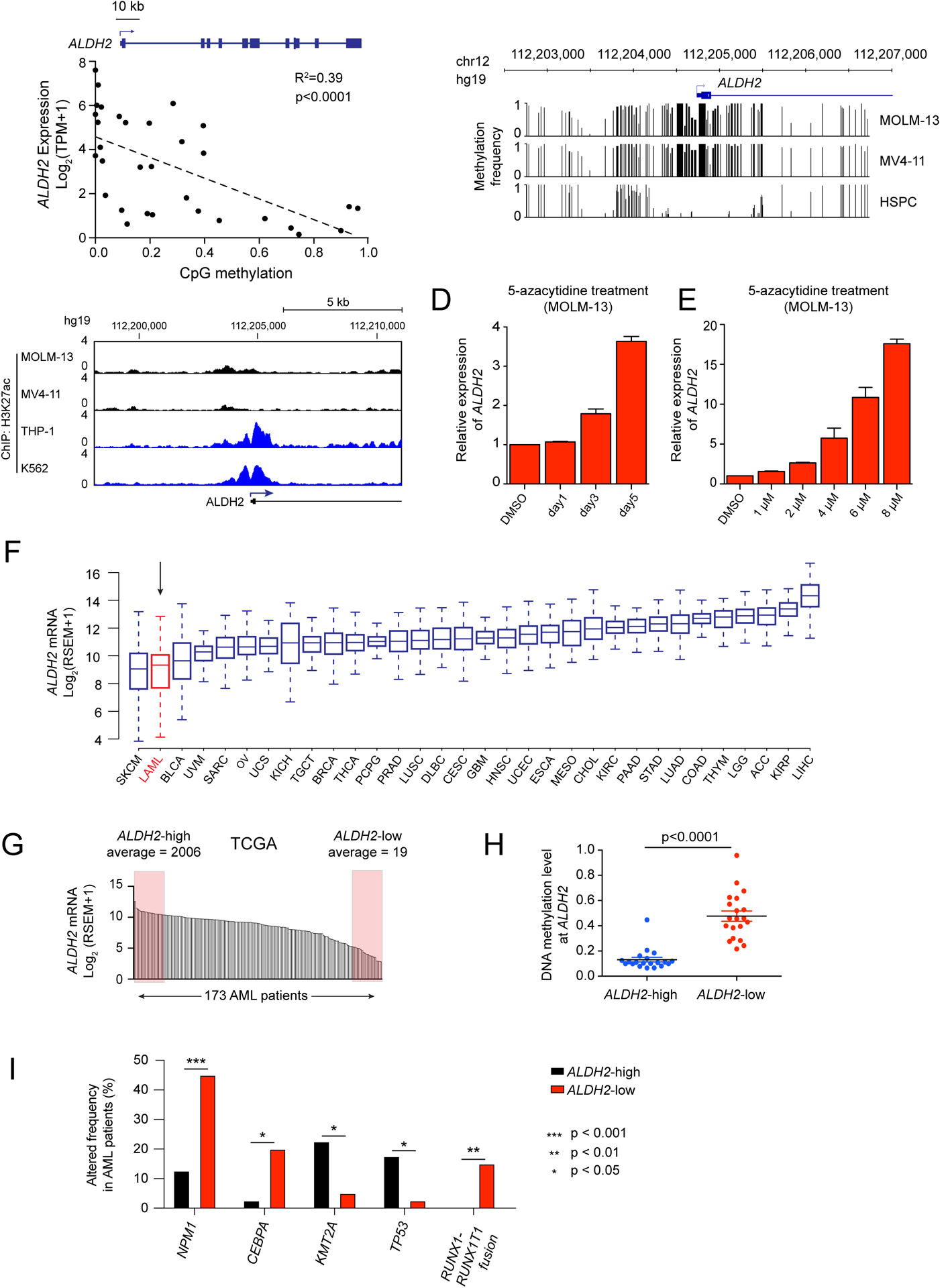
(A) Scatterplot analysis comparing ALDH2 mRNA levels and DNA methylation levels at the ALDH2 promoter across 32 AML cell lines (data was obtained from the Cancer Cell Line Encyclopedia) (28,40). p value is calculated by linear regression. (B) DNA methylation analysis of the ALDH2 promoter in leukemia cell lines (Nanopore sequencing) or in normal human hematopoietic stem and progenitor cells (bisulfite sequencing from (42)). (C) ChIP-seq analysis of H3K27ac enrichment at the ALDH2 promoter. (D-E) RT-qPCR analysis of ALDH2 mRNA levels in MOLM-13 cells following treatment with 5-azacytidine. In (D), 1 μM concentration was used. In (E), a 36 hour timepoint was used. (F) ALDH2 mRNA levels extracted from the TCGA Pan-Can Atlas (71). (G) ALDH2 mRNA levels in 173 AML patient samples from TCGA. Shown are the samples classified as ALDH2-high and ALDH2-low. (H) Comparison of CpG DNA methylation level at the ALDH2 promoter of indicated AML patient samples. p-value is calculated by unpaired Student’s t-test. (I) Genetic alterations that correlate with ALDH2 silencing in human AML patients. ALDH2-low and ALDH2-high patient samples were extracted from the TCGA Pan-Cancer database, and the genetic alteration frequencies were evaluated in these two groups of patients. An unpaired student’s t test was performed to determine statistical significance, which revealed the five genes shown as being significantly correlated or anti-correlated with ALDH2 silencing. All bar graphs represent the mean ± SEM (n=3).
