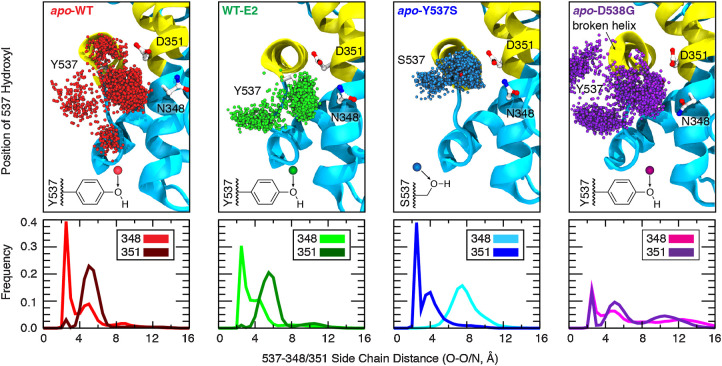
Figure 1.
Effect of ligand binding and mutation on hydrogen-bonding contacts. The dynamics of hydrogen-bonding partners Y/S537 (H12) with N348 and D351 (H3) were evaluated from replicate 200-ns simulations of dimeric ERα complexes (1.2 μs aggregate sampling for each system). Top, the position of the Y/S537 side-chain oxygen is shown every 200 ps (3,000 spheres; only monomer A is shown for clarity). Bottom, the shortest distance between side-chain heteroatoms of Y/S537(O) and N348(O/N) or D351(O/O) were binned (width = 0.5 Å) to yield frequency histograms of each interaction.