Figure 1. Transcriptional networks governed by RB are stage-dependent.
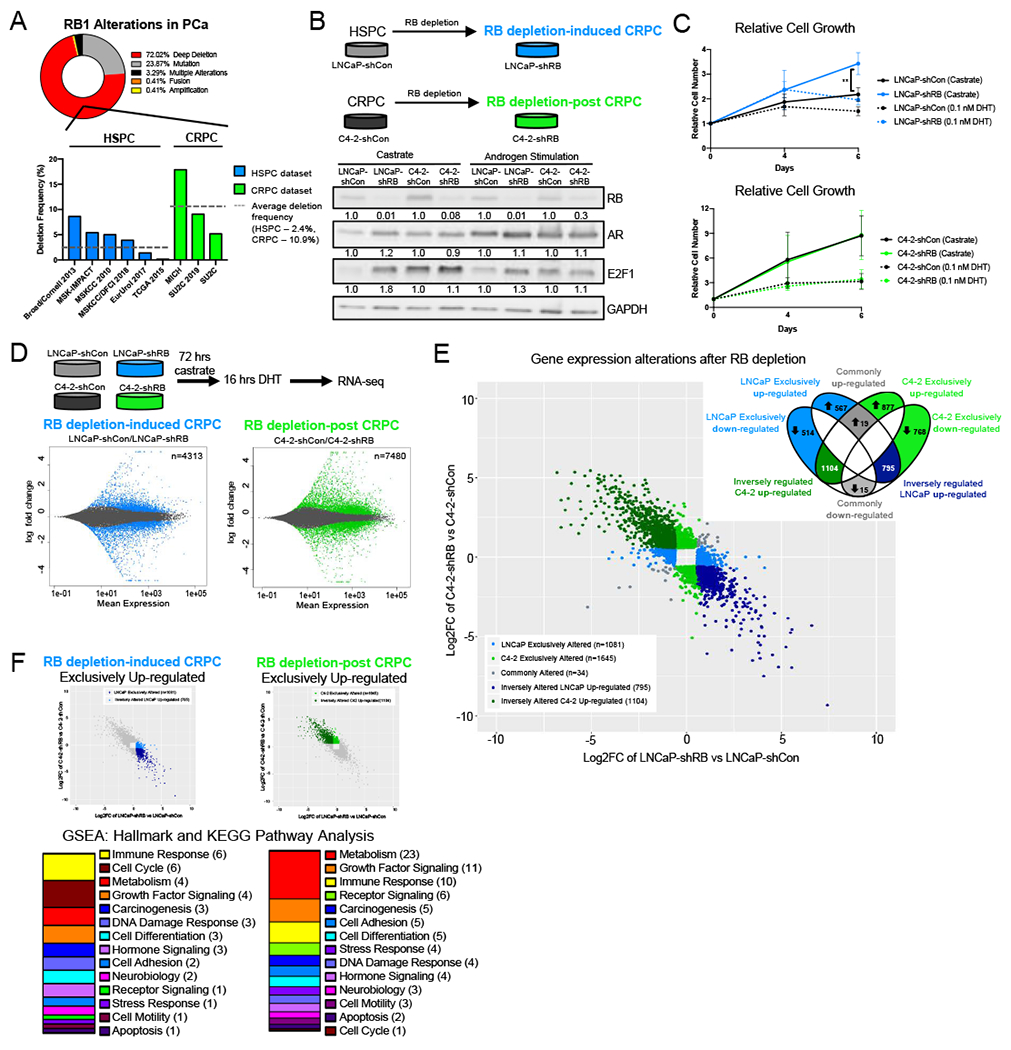
(A) RB1 alterations across prostate adenocarcinoma and CRPC datasets from CBioPortal. Frequency of RB1 gene deep across HSPC and CRPC cohorts. (B) RB1 was depleted via shRNA in LNCaP and C4-2 cell lines to generate RB1 depletion-induced CRPC (RBD-induced CRPC) and RB1 depletion-post CRPC models (RBD-post CRPC), respectively. Relative protein expression of RB, AR, and E2F1 in castrate and androgen stimulated (10 nM DHT) conditions are shown by western blot. (C) Relative growth of LNCaP-shCon, LNCaP-shRB, C4-2-shCon and C4-2-shRB models in castrate and androgen stimulated conditions (0.1 nM DHT). (D) RNA sequencing was performed in LNCaP and C4-2 isogenic models after 72 hours androgen depletion followed by 16 hrs of DHT stimulation. MA plot representing transcript alterations after RB knockdown from RNA-seq in RB depletion induced CRPC and RB depletion post CRPC models. Genes highlighted in blue (LNCaP) and green (C4-2) represent statistically significant differential expression (adjusted p-value < 0.05) (E) Transcripts altered in both models with a fold change >1.5 were plotted on the same graph. Transcripts were divided into those exclusively altered in either model after RB depletion (LNCaP 1081-blue, C4-2 1645-green) and those inversely regulated, upregulated in LNCaP and down regulated in C4-2 after RB depletion (795-dark blue) and downregulated in LNCaP and upregulated in C4-2 (1104-dark green). Transcripts commonly altered in both models are shown in grey (34). (F) GSEA Hallmark and KEGG pathway analysis from the molecular signatures database (MSigDB) of transcripts upregulated in the ONLY in the LNCaP-shRB model and pathway analysis of transcripts upregulated in the ONLY in the C4-2-shRB model. Pathways were categorized into more broad descriptions for comparison.
