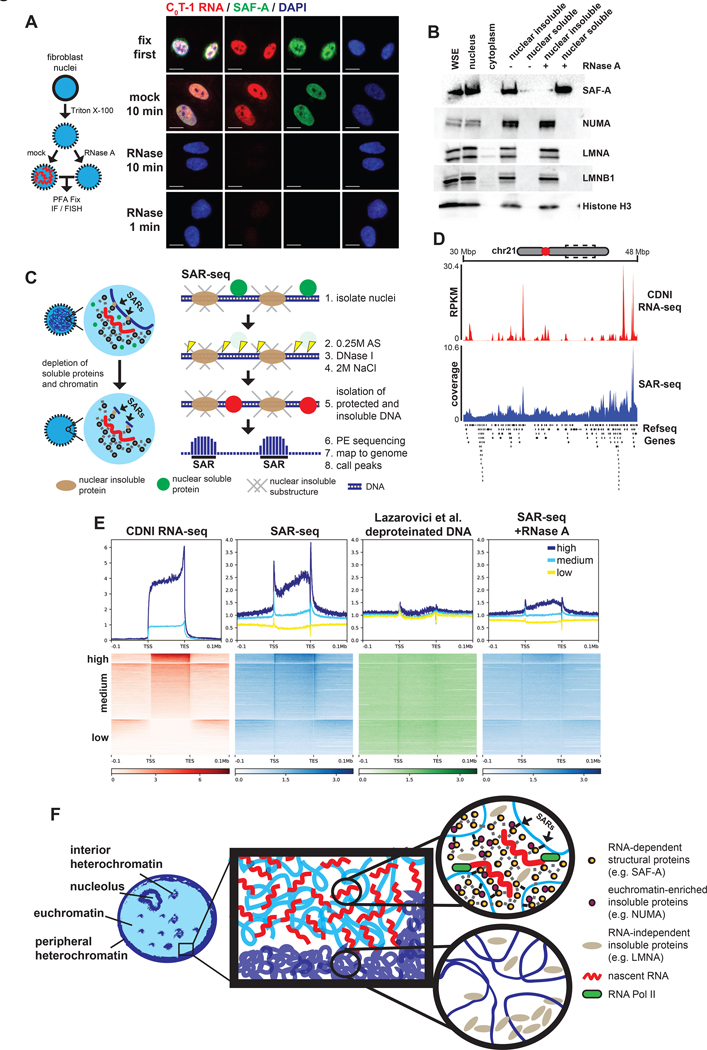
Figure 7. RNA is required for SAF-A to interact with chromatin and influences nuclear scaffold attachment.
(A) IF or RNA FISH analysis on permeabilized nuclei mock or RNase-treated for 10 minutes at 37°C prior to fixation. Images representative of >95% of nuclei. Scale bars 10 μm. (B) Western blot of protein solubility following gentle centrifugation of mock or RNase-treated nuclei. (C) Schematic of scaffold attachment region isolation and sequencing (SAR-seq) protocol. Soluble proteins and fragmented chromatin are extracted from isolated nuclei. Genomic DNA protected from digestion that remained insoluble after ionic extraction was subjected to paired-end sequencing without fixation or additional fragmentation. (D) Coverage map of CDNI RNA-seq (red) or SAR-seq (blue) at an 18 Mbp region of chr21. Refseq protein-coding genes annotated below. (E) Heatmap and average coverage profiles of CDNI RNA-seq (RPKM), SAR-seq (normalized coverage), highly DNase I digested naked (deproteinized) DNA-seq (normalized coverage), and SAR-seq from nuclei treated with RNase A for 10 minutes. Regions were binned by mean transcript abundance in CDNI and NS fractions. (F) Model summarizing results consistent with a dynamic ribonucleoprotein (RNP) meshwork, or scaffold, platformed on nascent transcripts that antagonizes cytologically observable chromatin compaction. RNP scaffold components (eg. SAF-A, MATR3, and NUMA) co-distribute with nascent RNA. In contrast, nascent RNA and RNP scaffold factors are depleted from regions of heterochromatin and experimentally compacted chromatin. Regions of chromatin near active transcription preferentially interact with the insoluble nuclear scaffold. Results here indicate RNA-dependent interactions between chromatin and the RNP-scaffold may serve to maintain decondensed chromatin structure.