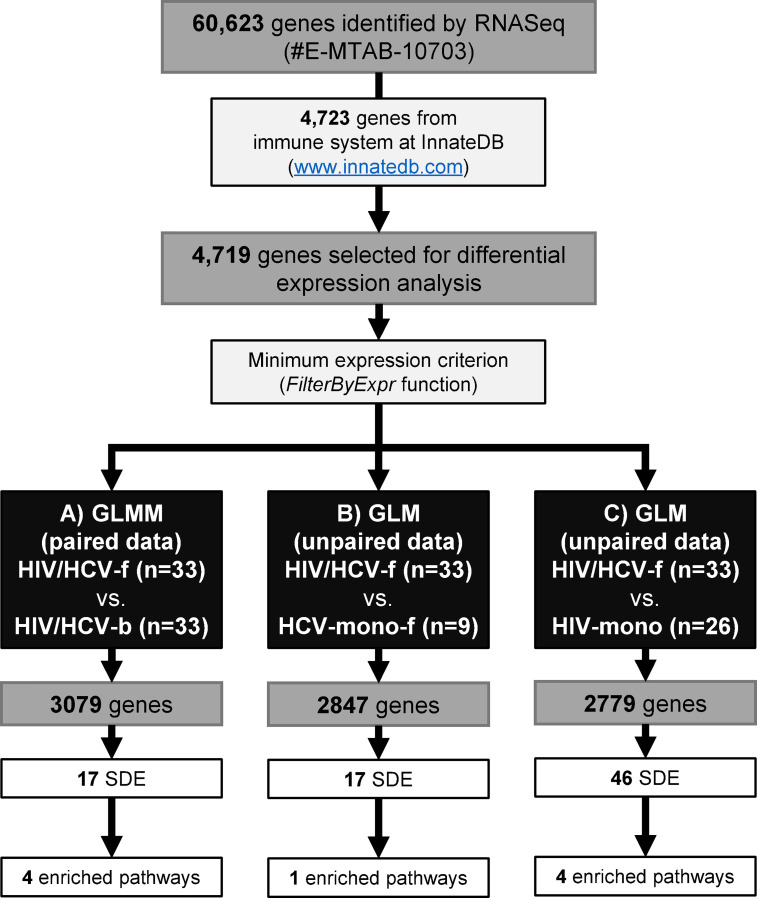
Figure 1.
Study design flowchart. RNA sequencing identified a total of 60,623 genes. Of these, we selected 4,723 genes of the immune system from the InnateDB. A total of 4,719 genes were selected for the differential expression analysis. Next, the FilterByExpr function was applied for each comparison: (A) HIV/HCV-f vs. HIV/HCV-b, 3,079 genes were selected, 17 were SDE genes involved in four biological pathways; (B) HIV/HCV-f vs. HCV-mono-f, 2,847 genes were selected, 17 were SDE genes involved in one biological pathway; (C) HIV/HCV-f vs. HIV-mono, 2,779 genes were selected, 46 were SDE genes involved in four biological pathways. HIV, human immunodeficiency virus; HCV, hepatitis C virus; HIV/HCV-f, HIV/HCV-coinfected patients 36 weeks after the sustained virological response (SVR); HCV-mono-f, HCV-monoinfected patients 36 weeks after SVR; HIV-mono, HIV-monoinfected patients; SDE, significantly differentially expressed; GLM, generalized linear model; GLMM, Generalized linear mixed model.