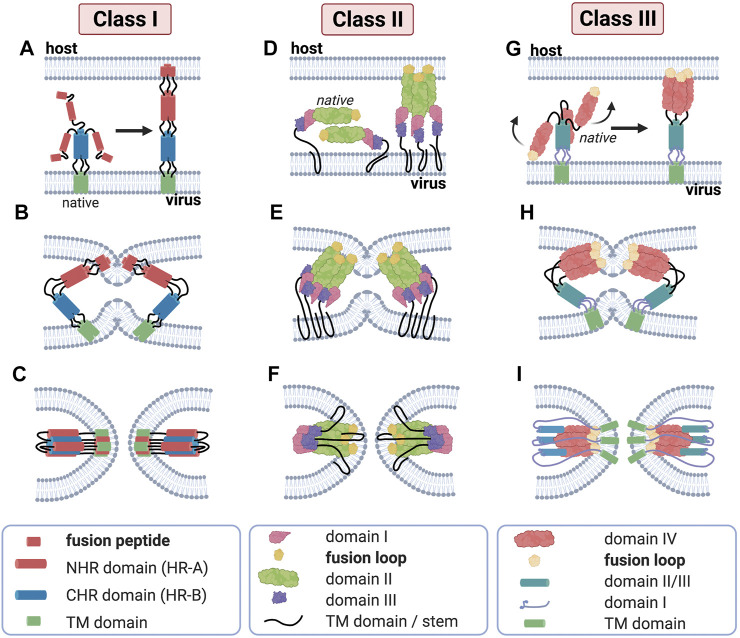
FIGURE 3.
Major structural features of membrane fusion processes across the three canonical classes of fusion protein. (Column 1) Membrane fusion driven by viral class I proteins. The model of class I protein-mediated hemifusion and fusion depicts the progression through an extended prehairpin followed by breaking the threefold symmetry and dissociation of the C-heptad repeat domains. In the native prefusion conformation, the paramyxovirus fusion (F) protein consists of globular head domain attached to the transmembrane (TM) domains and short luminal tails through the TM domain-proximal heptad repeat sequences. When fusion commences, major structural rearrangements lead to assembly of the head-domain HR segments into a central, trimeric alpha-helical coiled-coil structure, displacing the fusion peptides in the direction of the host-cell membrane (A). Subsequent hairpin-like refolding (B) then positions the heptad repeat domains into the grooves of the central triple helix, resulting in the formation of the stable six-helix bundle (6HB) post-fusion structure (C), in which the fusion peptide-proximal core coiled-coil structure is surrounded by the three TMD-proximal HR helices; this is a defining feature of fusion proteins of this class. (Column 2) Class II viral protein-mediated fusion. Homo-/hetero-di-/trimers of E1/E2 (D) engage a target membrane with their fusion loops following low pH-induced conformational changes. Subsequent conformational changes involve the reorientation of domain III around a hinge region (E) that positions the TM region and the fusion loop (yellow) proximal to one another. Subsequent refolding of the extended trimeric conformation into a hairpin structure promotes hemifusion and fusion pore formation (F). Despite their marked structural differences, proteins of class I and class II are able to progress through a similar refolding pathway. (Column 3) Membrane merger induced by class III viral fusogens. The native trimer of class III fusion proteins folds into a tripod-like arrangement, on which the fusion loops are positioned at the tip of each leg and are therefore directed into the viral envelope. Low pH conditions result in the protonation of a specific cluster of histidine residues that exert a major destabilizing effect on the prefusion structure. Once triggered, the tripod legs are proposed to swing upward (G), driving the fusion loops toward the target membrane. Repositioning of the domains with conserved tertiary structure relative to each other through secondary structure reorganization in hinge regions and major changes of the trimerization domain (H) then ultimately result in a classic hairpin post-fusion conformation (I).