Figure 1. APOBEC3A interacts with all subunits of the CCT chaperonin complex.
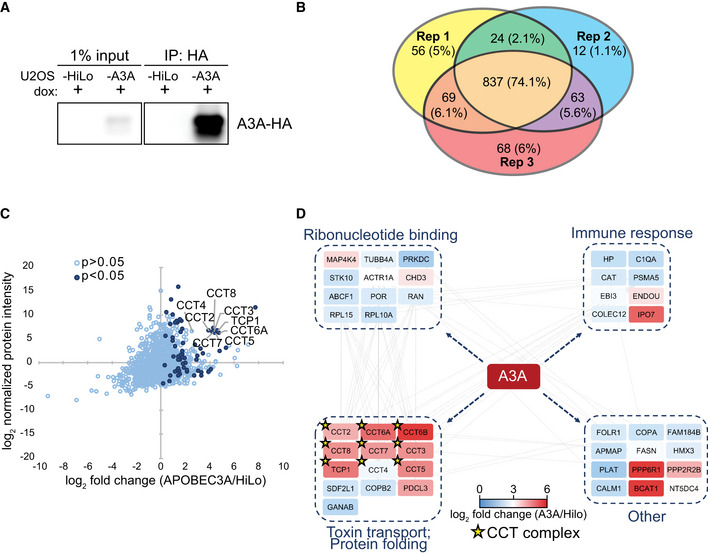
- Immunoprecipitation of A3A. U2OS‐HiLo cells with doxycycline‐inducible, haemagglutinin (HA)‐tagged A3A (U2OS‐A3A) and parental cells (U2OS‐HiLo without transgene) were treated with dox for 72 h. Immunoblot detection of A3A‐HA after immunoprecipitation (IP) of cell lysates with anti‐HA antibody is shown.
- Comparison of mass spectrometry results from three (n = 3) replicates. IP followed by mass spectrometry was performed in triplicate for parental U2OS‐HiLo and U2OS‐A3A cell lines. Venn diagram shows the overlap of proteins identified in three biological replicates of HA immunoprecipitation from U2OS‐A3A cells. Number of proteins identified in each replicate is displayed along with the percentage of protein overlap identified between replicates.
- All eight subunits of the CCT chaperonin complex were enriched in U2OS‐A3A samples. Proteins detected by MS are displayed by fold enrichment (log2) from U2OS‐A3A cells compared to parental cells (x‐axis) and by protein abundance within the U2OS‐A3A samples (y‐axis). Significantly enriched proteins (P < 0.05 by one‐tailed t‐test, n = 3) are denoted in dark blue. Components of the CCT complex are highlighted and labeled.
- Network of specific interactors of APOBEC3A. Only proteins significantly enriched in A3A samples over HiLo were considered for network analysis (log2 fold change > 0; one‐tailed t‐test P < 0.0.5), with the exception of CCT6B and CCT4 (t‐test P > 0.05). Node colors denote enrichment (indicated by heatmap legend) of interacting protein in A3A samples compared to HiLo. Nodes are grouped into boxes according to gene ontology enriched biological processes for interacting proteins (STRING database, FDR < 0.01). Gray edge lines indicate observed interactions between proteins. CCT complex members are denoted by a star.
Source data are available online for this figure.
