Figure 5. A3A mutational signatures are enriched in cancers with CCT mutations.
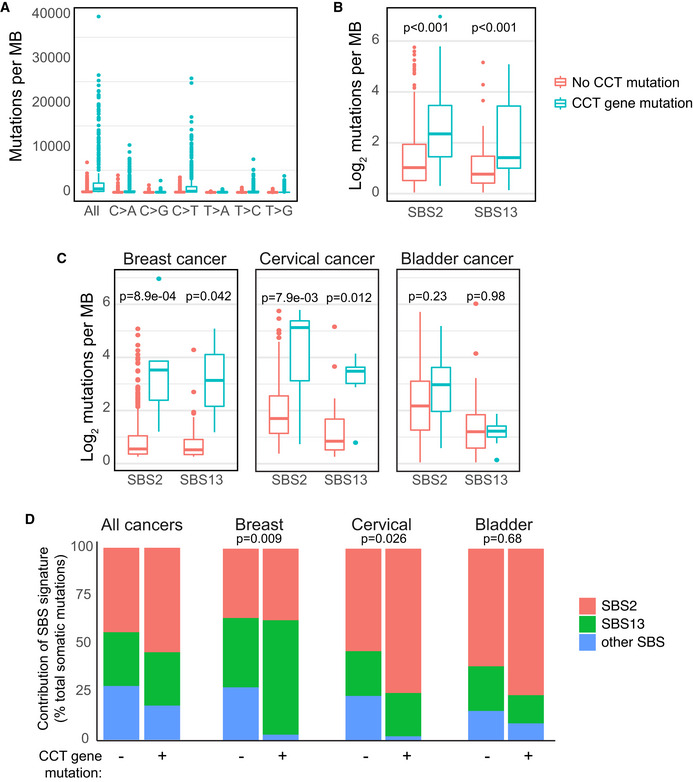
- Tumors with CCT gene mutations have an increased mutational burden. All cancer genome sequences in TCGA were evaluated for mutation burden and specific base substitutions. Cancers with CCT gene mutations predicted to cause a negative impact on protein function were classified as deleterious and were compared to those with no mutations in any CCT gene. Wilcoxon rank sum test applied to all paired comparisons between tumors with and without CCT gene mutations yielded P < 0.001.
- The APOBEC3 mutational signature is enriched in tumors with CCT gene mutations. Single base substitution (SBS) patterns, as defined by COSMIC Mutational Signatures v2, were evaluated in CCT‐mutated and CCT non‐mutated cancer genome sequences. Depicted are the mutational signatures attributable to APOBEC3 activity (SBS2 and SBS13). Denoted P‐values determined by Wilcoxon rank sum test.
- Quantification of APOBEC3 mutational signatures among three tumor types that have previously been associated with high APOBEC3 activity. Breast, bladder, and cervical cancer samples from the TCGA database were divided into CCT‐mutated and non‐mutated genomes, as above. The number of mutations contributing to SBS2 and SBS13 in each tumor type was quantified and displayed as an average. Comparison by Wilcoxon rank sum test yielded P‐values denoted.
- The contribution of APOBEC3 mutational signatures (SBS2 and SBS13) and all other SBS signatures defined by COSMIC v2 was evaluated in all tumors and independently in breast, bladder, and cervical cancer genomes within the TCGA database. A comparison of mutational signature contributions between CCT‐mutated and non‐mutated tumors is shown. A z‐test of proportions was used to compare whether the percentage of “other” signatures is significantly different between CCT‐mutated and non‐mutated tumors, and P‐values show statistical significance in breast and cervical cancers but not in bladder cancer.
Data information: Box and whisker plots depict median and interquartile range (25–75%).
