-
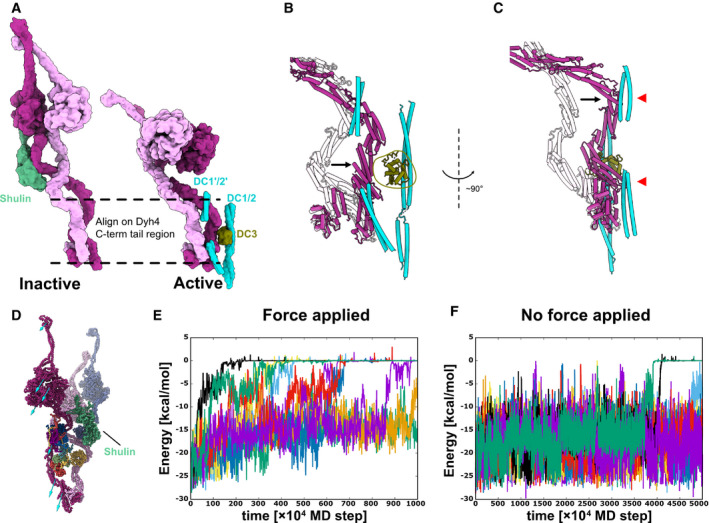
A
Models of the inactive and the active ODA. Dyh3 and Dyh4 parts from the models are shown with Shulin and docking complex, respectively. The two structures are aligned based on the Dyh4 residues 414–513 (helix bundle 3).
-
B, C
Detailed comparison of the Dyh3 before and after the remodeling. With the two structures aligned as in (A), Dyh3 tail in the active conformation indicates significant shift toward DC3 (B). Similarly, the Dyh3 tail in the active conformation indicates a significant shift toward the extended coiled‐coil region of DC1/2 (˜ 320–400 aa). Red arrowheads indicate the interaction sites of Dyh3 with the extended coiled‐coil region of DC1/2.
-
D
Coarse grain molecular dynamics of the inactive Shulin–ODA complex. External force was applied in the direction of the arrow to simulate the remodeling of Dyh3 upon binding to the doublet microtubule. Shulin is shown in green.
-
E, F
Energy trajectories between Shulin and the ODA complex with (E) and without (F) external force applied to Dyh3. 8 out of 10 setups showed detachment of Shulin (energy = 0) with external force in 1,000 × 104 MD steps, which roughly corresponds to 10 μs. (E). In contrast, only two out of 10 trajectories showed Shulin detachment without applied force even with 5,000 × 104 MD steps, which approximately corresponds to 50 μs. (F). It should be noted that our coarse‐grained MD simulation is performed in the condition where Shulin binding to the ODA complex is weakened, but it still showed the difference in detachment rate of Shulin in response to the global conformational change in the ODA complex.