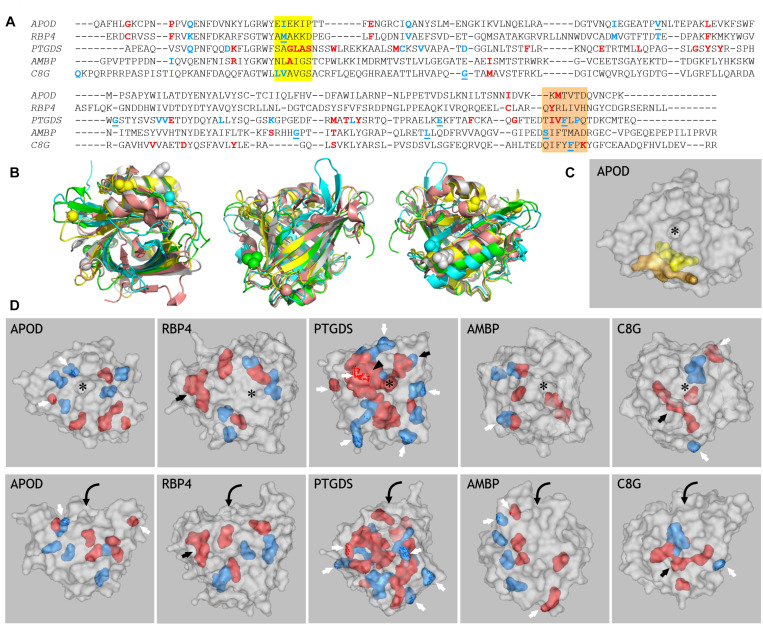
FIGURE 3.
Prediction of amino acid residues related to functional divergence in ED Lipocalins. (A) MSA of the five human representatives of the ED Lipocalins selected for this analysis. Residues related to Type-I divergence are labeled in blue, with underlined residues showing SSPP-probability support. Specificity-determining positions (SDPs) are labeled in red. Colored boxes mark two sequence regions enriched in functionally divergent residues. (B) Human Lipocalin structures superimposed, based on the structurally conserved beta-barrel, and colored (APOD, green; RBP4, cyan; PTGDS, gray; AMBP, yellow; C8G, salmon). Views into the ligand pocket, at the front side and the back side, revealing the alpha-helix attached to the beta-barrel, are shown. Colored spheres display SSPP-supported Type-I residues in all five Lipocalins. (C) Space-filled view of ApoD highlighting the residues marked by the two colored boxes in (A). (D) Gray-colored space-filled views of the five human Lipocalin structures showing the color-coded residues identified in (A). Panels in the upper row show views into the Lipocalin hydrophobic pocket (asterisk) while panels in the lower row show side views of the beta-barrel. Residues accessible at the protein surface (white arrows), or buried within the structure (black arrows) are pointed. Arrowhead points to a side wall of the binding pocket in PTGDS, and curved arrows suggest the binding pocket entrance on the side views of Lipocalins.