Figure 1.
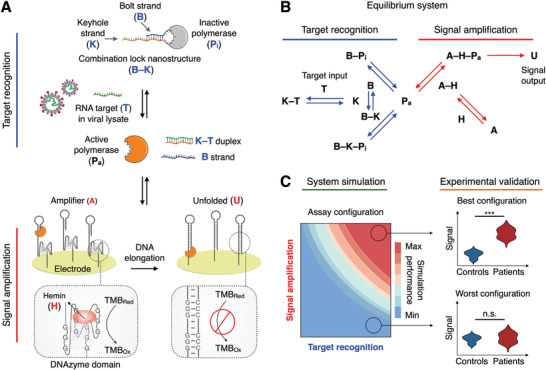
The SCREEN platform. A) Working principle of the SCREEN platform. SCREEN leverages the favorable coupling of two catalytic networks: the target recognition network and the signal amplification network. The target recognition network consists of a programmable DNA lock nanostructure (B–K) which binds and inactivates a DNA polymerase (Pi). The lock nanostructure comprises a bolt strand (B) and a keyhole strand (K) that is designed to be complementary to the target RNA (T). In the presence of the target, the keyhole strand hybridizes with the target, displacing the bolt strand; this displacement destabilizes the lock nanostructure and releases an active polymerase (Pa). Polymerase activation is next measured through the signal amplification network, which consists of amplifier DNA nanostructures (A) immobilized on the surface of a screen‐printed electrode. Each amplifier nanostructure comprises an upstream polymerase‐binding domain, a hairpin structure that primes any polymerase activity, and a downstream DNAzyme peroxidase domain, assembled through G‐quadruplex coupling with hemin (H). As an active polymerase binds and elongates the amplifier nanostructure, the polymerase disrupts the latter's G‐quadruplex peroxidase domain. The unfolded amplifier (U) thus loses its catalytic activity for redox reactions and produces a large resultant signal change. B) SCREEN network system. SCREEN leverages collaborative equilibrium coupling across multiple interactions within the network architecture to efficiently drive the dual‐enzyme catalysis, thereby transducing even sparse target input (T) into an amplified and rapid signal output (U). C) To establish favorable equilibrium shifts, we developed a computational model that simulated all reactions in the network system to tune molecular components, predict overall assay performance and establish equilibrium‐driven assay configurations. The simulation‐derived assay configurations were experimentally validated and clinically evaluated for COVID‐19 detection. n.s., not significant. *** indicates statistical significance.
