Figure 4.
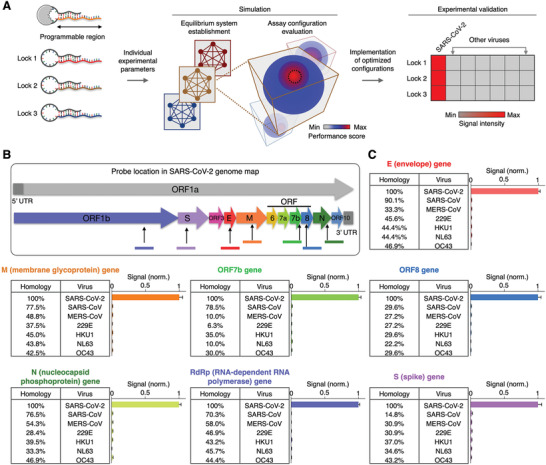
Programmable SCREEN assays for different targets. A) New SCREEN assay development. The lock nanostructure contains a programmable region that can be varied at will to detect different targets of interest. For these new lock designs, we experimentally characterize their respective equilibrium and kinetic parameters, and employ the SCREEN computation model to establish and evaluate the performance of millions of assay configurations. Optimized SCREEN assays are selected from the simulation study and experimentally validated. B) SARS‐CoV‐2 genome map. Gene sections of interest used in this study are indicated. Not drawn to scale. C) SCREEN assays for seven different SARS‐CoV‐2 gene targets. Using the SCREEN assay development workflow, we established new assays for different SARS‐CoV‐2 gene targets and experimentally evaluated their specificity, through testing with other pathogen sequences with various levels of homology. All measurements were performed in triplicate and the data are presented as mean± s.d.
