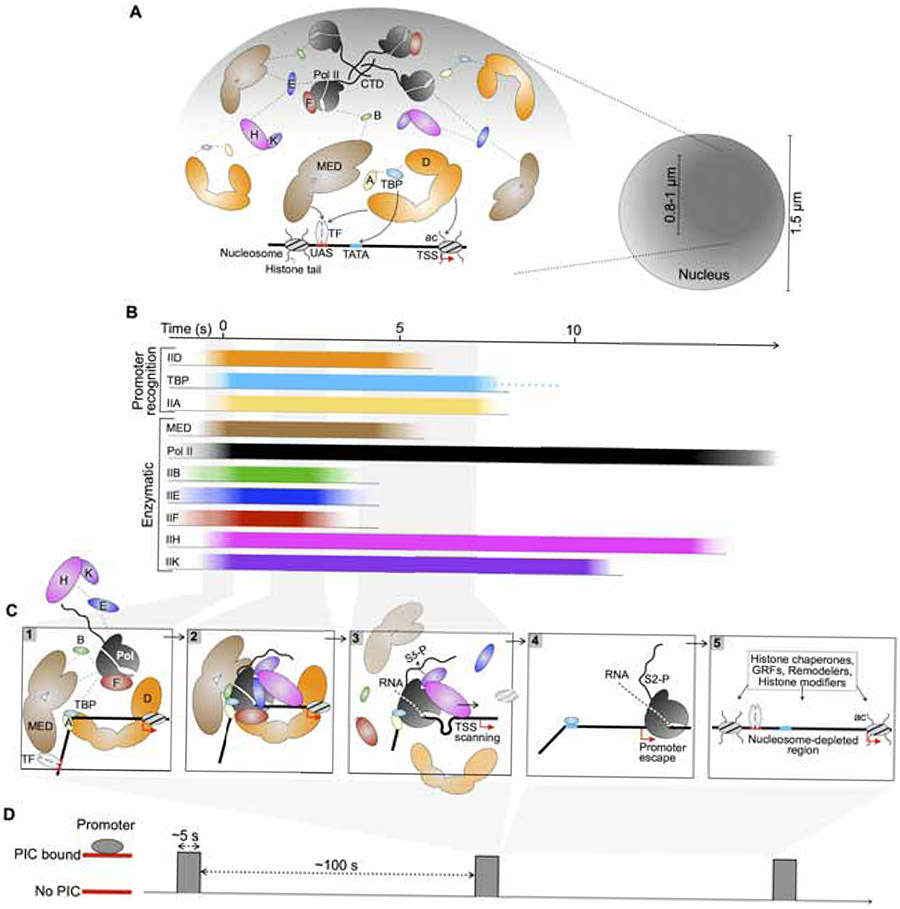
Figure 7. Spatio-temporal model of PIC establishment in vivo.
(A) Left: A model for spatial clustering supported by multi-valent interactions (dashed lines) among promoter-recognizing PIC components TFIID, TBP and TFIIA as well as Mediator and enzymatic components. The presence of active genes in this region is implicated, with one stereotypical promoter is shown. Mediator and TFIID may interact with DNA elements, bound sequence-specific transcription factor (TF), and the +1 nucleosome via acetylated histone tails (ac). Right: Schematic of a subnuclear environment (dark shade) explored by PIC components in a subdiffusive manner. The diameter of this region was estimated based on median Rc values (0.4-0.5 μm) (Figure 1F).
(B) An average timescale of association and dissociation of PIC components at a yeast promoter. Recruited components are shown to associate in a quasi-synchronous manner based on calculated kobs (Figure 5G). Binding events are temporally scaled according to average τsb (Figure 3C) and estimated for TBP and Pol II.
(C) Sequential stages of PIC assembly and disassembly according to binding and dissociation of individual components in (B).
(D) An average timescale depicting sparse PIC establishment at a promoter, based on the ~5 s assembly window outlined in (B) and the association rate kobs = 0.01 s−1 estimated for Mediator and TFIID (Figure 5G).
See also Figure S8.